Reading and erasing of the phosphonium analogue of trimethyllysine by epigenetic proteins
- PMID: 36071790
- PMCID: PMC7613515
- DOI: 10.1038/s42004-022-00640-4
Reading and erasing of the phosphonium analogue of trimethyllysine by epigenetic proteins
Abstract
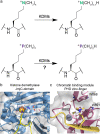
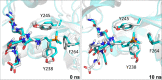
N ε-Methylation of lysine residues in histones plays an essential role in the regulation of eukaryotic transcription. The 'highest' methylation mark, N ε-trimethyllysine, is specifically recognised by N ε-trimethyllysine binding 'reader' domains, and undergoes demethylation, as catalysed by 2-oxoglutarate dependent JmjC oxygenases. We report studies on the recognition of the closest positively charged N ε-trimethyllysine analogue, i.e. its trimethylphosphonium derivative (KPme3), by N ε-trimethyllysine histone binding proteins and Nε-trimethyllysine demethylases. Calorimetric and computational studies with histone binding proteins reveal that H3KP4me3 binds more tightly than the natural H3K4me3 substrate, though the relative differences in binding affinity vary. Studies with JmjC demethylases show that some, but not all, of them can accept the phosphonium analogue of their natural substrates and that the methylation state selectivity can be changed by substitution of nitrogen for phosphorus. The combined results reveal that very subtle changes, e.g. substitution of nitrogen for phosphorus, can substantially affect interactions between ligand and reader domains / demethylases, knowledge that we hope will inspire the development of highly selective small molecules modulating their activity.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources

