HupB, a nucleoid-associated protein, is critical for survival of Mycobacterium tuberculosis under host-mediated stresses and for enhanced tolerance to key first-line antibiotics
- PMID: 36071978
- PMCID: PMC9441915
- DOI: 10.3389/fmicb.2022.937970
HupB, a nucleoid-associated protein, is critical for survival of Mycobacterium tuberculosis under host-mediated stresses and for enhanced tolerance to key first-line antibiotics
Abstract
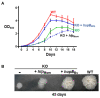
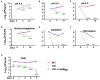
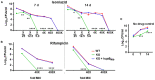
To survive and establish its niche, Mycobacterium tuberculosis (Mtb) engages in a steady battle against an array of host defenses and a barrage of antibiotics. Here, we demonstrate that Mtb employs HupB, a nucleoid-associated protein (NAP) as its key player to simultaneously battle and survive in these two stress-inducing fronts. Typically, NAPs are key to bacterial survival under a wide array of environmental or host-mediated stresses. Here, we report that for Mtb to survive under different macrophage-induced assaults including acidic pH, nutrient depletion, oxidative and nitrosative stresses, HupB presence is critical. As expected, the hupB knockout mutant is highly sensitive to these host-mediated stresses. Furthermore, Mtb aptly modulates HupB protein levels to overcome these stresses. We also report that HupB aids Mtb to gain tolerance to high levels of rifampicin (RIF) and isoniazid (INH) exposure. Loss of hupB makes Mtb highly susceptible to even short exposures to reduced amounts of RIF and INH. Overexpressing hupB in Mtb or complementing hupB in the hupB knockout mutant triggers enhanced survival of Mtb under these stresses. We also find that upon loss of hupB, Mtb significantly enhances the permeability of its cell wall by modulating the levels of several surface lipids including phthiocerol dimycocerosates (PDIMs), thus possibly influencing overall susceptibility to host-mediated stresses. Loss of hupB also downregulates efflux pump expression possibly influencing increased susceptibility to INH and RIF. Finally, we find that therapeutic targeting of HupB with SD1, a known small molecule inhibitor, significantly enhances Mtb susceptibility to INH and THP-1 macrophages and significantly reduces MIC to INH. Thus, our data strongly indicate that HupB is a highly promising therapeutic target especially for potential combinatorial shortened therapy with reduced INH and RIF doses.
Keywords: Hlp; HupB; Mycobacterium smegmatis; Mycobacterium tuberculosis; SD1; antibiotics; stress.
Copyright © 2022 Singh, Sharma, Singh, Pandey, Ilyas, Sisodiya, Choudhury, Gosain, Singh and Atmakuri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

