Epigenetic regulation of transcription factor binding motifs promotes Th1 response in Chagas disease cardiomyopathy
- PMID: 36072583
- PMCID: PMC9441916
- DOI: 10.3389/fimmu.2022.958200
Epigenetic regulation of transcription factor binding motifs promotes Th1 response in Chagas disease cardiomyopathy
Abstract
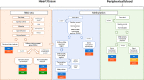
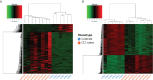
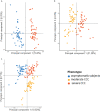
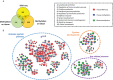
Chagas disease, caused by the protozoan Trypanosoma cruzi, is an endemic parasitic disease of Latin America, affecting 7 million people. Although most patients are asymptomatic, 30% develop complications, including the often-fatal Chronic Chagasic Cardiomyopathy (CCC). Although previous studies have demonstrated some genetic deregulations associated with CCCs, the causes of their deregulations remain poorly described. Based on bulk RNA-seq and whole genome DNA methylation data, we investigated the genetic and epigenetic deregulations present in the moderate and severe stages of CCC. Analysis of heart tissue gene expression profile allowed us to identify 1407 differentially expressed transcripts (DEGs) specific from CCC patients. A tissue DNA methylation analysis done on the same tissue has permitted the identification of 92 regulatory Differentially Methylated Regions (DMR) localized in the promoter of DEGs. An in-depth study of the transcription factors binding sites (TFBS) in the DMRs corroborated the importance of TFBS's DNA methylation for gene expression in CCC myocardium. TBX21, RUNX3 and EBF1 are the transcription factors whose binding motif appears to be affected by DNA methylation in the largest number of genes. By combining both transcriptomic and methylomic analysis on heart tissue, and methylomic analysis on blood, 4 biological processes affected by severe CCC have been identified, including immune response, ion transport, cardiac muscle processes and nervous system. An additional study on blood methylation of moderate CCC samples put forward the importance of ion transport and nervous system in the development of the disease.
Keywords: Chagas disease; Th1 response; dilated cardiomyopathy; epigenetic; methylation; transcription factors.
Copyright © 2022 Brochet, Ianni, Laugier, Frade, Silva Nunes, Teixeira, Mady, Ferreira, Ferré, Santos, Kuramoto, Cabantous, Steffen, Stolf, Pomerantzeff, Fiorelli, Bocchi, Pissetti, Saba, Cândido, Dias, Sampaio, Gaiotto, Marin-Neto, Fragata, Zaniratto, Siqueira, Peixoto, Rigaud, Bacal, Buck, Almeida, Lin-Wang, Schmidt, Martinelli, Hirata, Donadi, Costa Pereira, Rodrigues Junior, Puthier, Kalil, Spinelli, Cunha-Neto and Chevillard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Cunha-Neto E, Dzau VJ, Allen PD, Stamatiou D, Benvenutti L, Higuchi ML, et al. . Cardiac gene expression profiling provides evidence for cytokinopathy as a molecular mechanism in chagas’ disease cardiomyopathy. Am J Pathol (2005) 167(2):305−13. doi: 10.1016/S0002-9440(10)62976-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

