Molecular physiology of Arc/Arg3.1: The oligomeric state hypothesis of synaptic plasticity
- PMID: 36073248
- PMCID: PMC9787330
- DOI: 10.1111/apha.13886
Molecular physiology of Arc/Arg3.1: The oligomeric state hypothesis of synaptic plasticity
Abstract
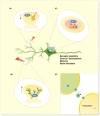
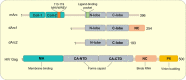
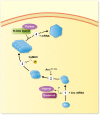
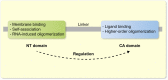
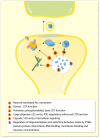
The immediate early gene, Arc, is a pivotal regulator of synaptic plasticity, memory, and cognitive flexibility. But what is Arc protein? How does it work? Inside the neuron, Arc is a protein interaction hub and dynamic regulator of intra-cellular signaling in synaptic plasticity. In remarkable contrast, Arc can also self-assemble into retrovirus-like capsids that are released in extracellular vesicles and capable of intercellular transfer of RNA. Elucidation of the molecular basis of Arc hub and capsid functions, and the relationship between them, is vital for progress. Here, we discuss recent findings on Arc structure-function and regulation of oligomerization that are giving insight into the molecular physiology of Arc. The unique features of mammalian Arc are emphasized, while drawing comparisons with Drosophila Arc and retroviral Gag. The Arc N-terminal domain, found only in mammals, is proposed to play a key role in regulating Arc hub signaling, oligomerization, and formation of capsids. Bringing together several lines of evidence, we hypothesize that Arc function in synaptic plasticity-long-term potentiation (LTP) and long-term depression (LTD)-are dictated by different oligomeric forms of Arc. Specifically, monomer/dimer function in LTP, tetramer function in basic LTD, and 32-unit oligomer function in enhanced LTD. The role of mammalian Arc capsids is unclear but likely depends on the cross-section of captured neuronal activity-induced RNAs. As the functional states of Arc are revealed, it may be possible to selectively manipulate specific forms of Arc-dependent plasticity and intercellular communication involved in brain function and dysfunction.
Keywords: Gag protein; activity-regulated cytoskeleton-associated protein (Arc); memory and cognition; oligomerization; protein interaction hub; protein structure-function; retrovirus-like capsid; synaptic plasticity.
© 2022 The Authors. Acta Physiologica published by John Wiley & Sons Ltd on behalf of Scandinavian Physiological Society.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

References
-
- Håvik B, Røkke H, Dagyte G, Stavrum A‐K, Bramham CR, Steen VM. Synaptic activity‐induced global gene expression patterns in the dentate gyrus of adult behaving rats: induction of immunity‐linked genes. Neuroscience. 2007;148(4):925‐936. - PubMed
-
- Bramham CR, Jensen KB, Proud CG. Tuning specific translation in cancer metastasis and synaptic memory: control at the MNK – eIF4E axis. Trends Biochem Sci. 2016;41(10):847‐858. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

