Targeting dual oncogenic machineries driven by TAL1 and PI3K-AKT pathways in T-cell acute lymphoblastic leukemia
- PMID: 36073513
- PMCID: PMC9890034
- DOI: 10.3324/haematol.2022.280761
Targeting dual oncogenic machineries driven by TAL1 and PI3K-AKT pathways in T-cell acute lymphoblastic leukemia
Abstract
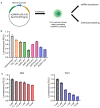
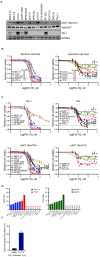
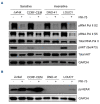
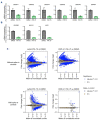
T-cell acute lymphoblastic leukemia (T-ALL) is a malignancy of thymic T-cell precursors. Overexpression of oncogenic transcription factor TAL1 is observed in 40-60% of human T-ALL cases, frequently together with activation of the NOTCH1 and PI3K-AKT pathways. In this study, we performed chemical screening to identify small molecules that can inhibit the enhancer activity driven by TAL1 using the GIMAP enhancer reporter system. Among approximately 3,000 compounds, PIK- 75, a known inhibitor of PI3K and CDK, was found to strongly inhibit the enhancer activity. Mechanistic analysis demonstrated that PIK-75 blocks transcriptional activity, which primarily affects TAL1 target genes as well as AKT activity. TAL1-positive, AKT-activated T-ALL cells were very sensitive to PIK-75, as evidenced by growth inhibition and apoptosis induction, while T-ALL cells that exhibited activation of the JAK-STAT pathway were insensitive to this drug. Together, our study demonstrates a strategy targeting two types of core machineries mediated by oncogenic transcription factors and signaling pathways in T-ALL.
Figures




References
-
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278(5340):1059-1064. - PubMed
-
- Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23(26):6306-6315. - PubMed
-
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8(5):380-390. - PubMed
-
- Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JP. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br J Haematol. 2008;143(2):153-168. - PubMed
-
- Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16(8):494-507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

