Activation of long non-coding RNA NEAT1 leads to survival advantage of multiple myeloma cells by supporting a positive regulatory loop with DNA repair proteins
- PMID: 36073514
- PMCID: PMC9827177
- DOI: 10.3324/haematol.2022.281167
Activation of long non-coding RNA NEAT1 leads to survival advantage of multiple myeloma cells by supporting a positive regulatory loop with DNA repair proteins
Abstract
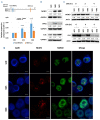
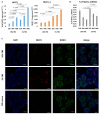
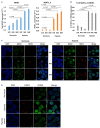
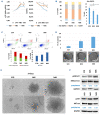
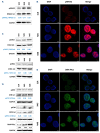
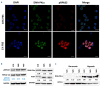
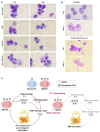
Long non-coding RNA NEAT1 is the core structural component of the nuclear paraspeckle (PS) organelles and it has been found to be deregulated in multiple myeloma (MM) patients. Experimental evidence indicated that NEAT1 silencing negatively impacts proliferation and viability of MM cells, both in vitro and in vivo, suggesting a role in DNA damage repair (DDR). In order to elucidate the biological and molecular relevance of NEAT1 upregulation in MM disease we exploited the CRISPR/Cas9 synergistic activation mediator genome editing system to engineer the AMO-1 MM cell line and generate two clones that para-physiologically transactivate NEAT1 at different levels. NEAT1 overexpression is associated with oncogenic and prosurvival advantages in MM cells exposed to nutrient starvation or a hypoxic microenvironment, which are stressful conditions often associated with more aggressive disease phases. Furthermore, we highlighted the NEAT1 involvement in virtually all DDR processes through, at least, two different mechanisms. On one side NEAT1 positively regulates the posttranslational stabilization of essential PS proteins, which are involved in almost all DDR systems, thus increasing their availability within cells. On the other hand, NEAT1 plays a crucial role as a major regulator of a molecular axis that includes ATM and the catalytic subunit of DNA-PK kinase proteins, and their direct targets pRPA32 and pCHK2. Overall, we provided novel important insightsthe role of NEAT1 in supporting MM cells adaptation to stressful conditions by improving the maintenance of DNA integrity. Taken together, our results suggest that NEAT1, and probably PS organelles, could represent a potential therapeutic target for MM treatment.
Figures

References
-
- Huarte M. The emerging role of lncRNAs in cancer. Nature Med. 2015;21(11):1253-1261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

