Constructing functional models from biophysically-detailed neurons
- PMID: 36074765
- PMCID: PMC9455888
- DOI: 10.1371/journal.pcbi.1010461
Constructing functional models from biophysically-detailed neurons
Abstract
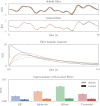
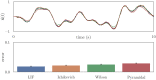
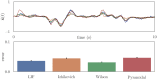
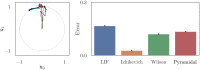
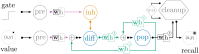
Improving biological plausibility and functional capacity are two important goals for brain models that connect low-level neural details to high-level behavioral phenomena. We develop a method called "oracle-supervised Neural Engineering Framework" (osNEF) to train biologically-detailed spiking neural networks that realize a variety of cognitively-relevant dynamical systems. Specifically, we train networks to perform computations that are commonly found in cognitive systems (communication, multiplication, harmonic oscillation, and gated working memory) using four distinct neuron models (leaky-integrate-and-fire neurons, Izhikevich neurons, 4-dimensional nonlinear point neurons, and 4-compartment, 6-ion-channel layer-V pyramidal cell reconstructions) connected with various synaptic models (current-based synapses, conductance-based synapses, and voltage-gated synapses). We show that osNEF networks exhibit the target dynamics by accounting for nonlinearities present within the neuron models: performance is comparable across all four systems and all four neuron models, with variance proportional to task and neuron model complexity. We also apply osNEF to build a model of working memory that performs a delayed response task using a combination of pyramidal cells and inhibitory interneurons connected with NMDA and GABA synapses. The baseline performance and forgetting rate of the model are consistent with animal data from delayed match-to-sample tasks (DMTST): we observe a baseline performance of 95% and exponential forgetting with time constant τ = 8.5s, while a recent meta-analysis of DMTST performance across species observed baseline performances of 58 - 99% and exponential forgetting with time constants of τ = 2.4 - 71s. These results demonstrate that osNEF can train functional brain models using biologically-detailed components and open new avenues for investigating the relationship between biophysical mechanisms and functional capabilities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures









References
-
- Anderson JR, Lebiere CJ. The atomic components of thought. Psychology Press; 2014.
-
- Eliasmith C, Anderson CH. Neural engineering: Computation, representation, and dynamics in neurobiological systems. MIT press; 2004.
-
- Eliasmith C. How to build a brain: A neural architecture for biological cognition. Oxford University Press; 2013.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

