The human disease gene LYSET is essential for lysosomal enzyme transport and viral infection
- PMID: 36074821
- PMCID: PMC9547973
- DOI: 10.1126/science.abn5648
The human disease gene LYSET is essential for lysosomal enzyme transport and viral infection
Abstract
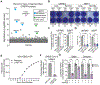
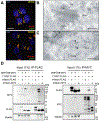
Lysosomes are key degradative compartments of the cell. Transport to lysosomes relies on GlcNAc-1-phosphotransferase-mediated tagging of soluble enzymes with mannose 6-phosphate (M6P). GlcNAc-1-phosphotransferase deficiency leads to the severe lysosomal storage disorder mucolipidosis II (MLII). Several viruses require lysosomal cathepsins to cleave structural proteins and thus depend on functional GlcNAc-1-phosphotransferase. We used genome-scale CRISPR screens to identify lysosomal enzyme trafficking factor (LYSET, also named TMEM251) as essential for infection by cathepsin-dependent viruses including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). LYSET deficiency resulted in global loss of M6P tagging and mislocalization of GlcNAc-1-phosphotransferase from the Golgi complex to lysosomes. Lyset knockout mice exhibited MLII-like phenotypes, and human pathogenic LYSET alleles failed to restore lysosomal sorting defects. Thus, LYSET is required for correct functioning of the M6P trafficking machinery and mutations in LYSET can explain the phenotype of the associated disorder.
Conflict of interest statement
Figures



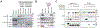
References
-
- Mercer J, Schelhaas M, Helenius A, Virus entry by endocytosis. Annual review of biochemistry 79, 803–833 (2010). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

