Pharmacogenomics polygenic risk score for drug response prediction using PRS-PGx methods
- PMID: 36075892
- PMCID: PMC9458667
- DOI: 10.1038/s41467-022-32407-9
Pharmacogenomics polygenic risk score for drug response prediction using PRS-PGx methods
Abstract
Polygenic risk scores (PRS) have been successfully developed for the prediction of human diseases and complex traits in the past years. For drug response prediction in randomized clinical trials, a common practice is to apply PRS built from a disease genome-wide association study (GWAS) directly to a corresponding pharmacogenomics (PGx) setting. Here, we show that such an approach relies on stringent assumptions about the prognostic and predictive effects of the selected genetic variants. We propose a shift from disease PRS to PGx PRS approaches by simultaneously modeling both the prognostic and predictive effects and further make this shift possible by developing a series of PRS-PGx methods, including a novel Bayesian regression approach (PRS-PGx-Bayes). Simulation studies show that PRS-PGx methods generally outperform the disease PRS methods and PRS-PGx-Bayes is superior to all other PRS-PGx methods. We further apply the PRS-PGx methods to PGx GWAS data from a large cardiovascular randomized clinical trial (IMPROVE-IT) to predict treatment related LDL cholesterol reduction. The results demonstrate substantial improvement of PRS-PGx-Bayes in both prediction accuracy and the capability of capturing the treatment-specific predictive effects while compared with the disease PRS approaches.
© 2022. Merck & Co., Inc., Rahway, NJ, USA and its affiliates.
Conflict of interest statement
S.Z., H.Z., D.V.M., and J.S. are employees at Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Rahway, NJ, USA.
Figures
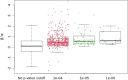
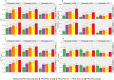
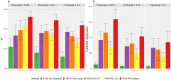
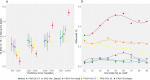
References
-
- Chen JJ, Lin WJ, Chen HC. Pharmacogenomic biomarkers for personalized medicine. Pharmacogenomics. 2013;14:969–980. - PubMed
-
- Roden DM, George Jr AL. The genetic basis of variability in drug responses. Nat. Rev. Drug Discov. 2002;1:37–44. - PubMed
-
- Nelson MR, et al. The genetics of drug efficacy: opportunities and challenges. Nat. Rev. Genet. 2016;17:197–206. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

