Heat-shock chaperone HSPB1 regulates cytoplasmic TDP-43 phase separation and liquid-to-gel transition
- PMID: 36075972
- PMCID: PMC9872726
- DOI: 10.1038/s41556-022-00988-8
Heat-shock chaperone HSPB1 regulates cytoplasmic TDP-43 phase separation and liquid-to-gel transition
Abstract
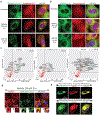
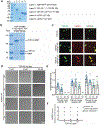
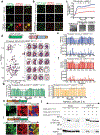
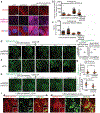
While acetylated, RNA-binding-deficient TDP-43 reversibly phase separates within nuclei into complex droplets (anisosomes) comprised of TDP-43-containing liquid outer shells and liquid centres of HSP70-family chaperones, cytoplasmic aggregates of TDP-43 are hallmarks of multiple neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS). Here we show that transient oxidative stress, proteasome inhibition or inhibition of the ATP-dependent chaperone activity of HSP70 provokes reversible cytoplasmic TDP-43 de-mixing and transition from liquid to gel/solid, independently of RNA binding or stress granules. Isotope labelling mass spectrometry was used to identify that phase-separated cytoplasmic TDP-43 is bound by the small heat-shock protein HSPB1. Binding is direct, mediated through TDP-43's RNA binding and low-complexity domains. HSPB1 partitions into TDP-43 droplets, inhibits TDP-43 assembly into fibrils, and is essential for disassembly of stress-induced TDP-43 droplets. A decrease in HSPB1 promotes cytoplasmic TDP-43 de-mixing and mislocalization. HSPB1 depletion was identified in spinal motor neurons of patients with ALS containing aggregated TDP-43. These findings identify HSPB1 to be a regulator of cytoplasmic TDP-43 phase separation and aggregation.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures






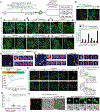


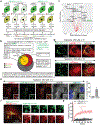

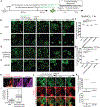


Comment in
-
Finding a chaperone for TDP-43.Nat Cell Biol. 2022 Sep;24(9):1328-1330. doi: 10.1038/s41556-022-00987-9. Nat Cell Biol. 2022. PMID: 36075973 No abstract available.
References
-
- Neumann M et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). - PubMed
-
- Neumann M et al. Absence of heterogeneous nuclear ribonucleoproteins and survival motor neuron protein in TDP-43 positive inclusions in frontotemporal lobar degeneration. Acta neuropathologica 113, 543–548 (2007). - PubMed
Method-only References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

