Event-triggered STED imaging
- PMID: 36076037
- PMCID: PMC9550628
- DOI: 10.1038/s41592-022-01588-y
Event-triggered STED imaging
Abstract
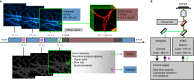
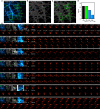
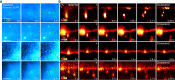
Monitoring the proteins and lipids that mediate all cellular processes requires imaging methods with increased spatial and temporal resolution. STED (stimulated emission depletion) nanoscopy enables fast imaging of nanoscale structures in living cells but is limited by photobleaching. Here, we present event-triggered STED, an automated multiscale method capable of rapidly initiating two-dimensional (2D) and 3D STED imaging after detecting cellular events such as protein recruitment, vesicle trafficking and second messengers activity using biosensors. STED is applied in the vicinity of detected events to maximize the temporal resolution. We imaged synaptic vesicle dynamics at up to 24 Hz, 40 ms after local calcium activity; endocytosis and exocytosis events at up to 11 Hz, 40 ms after local protein recruitment or pH changes; and the interaction between endosomal vesicles at up to 3 Hz, 70 ms after approaching one another. Event-triggered STED extends the capabilities of live nanoscale imaging, enabling novel biological observations in real time.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Alvelid J, Testa I. Fluorescence microscopy at the molecular scale. Curr. Opin. Biomed. Eng. 2019;12:34–42. doi: 10.1016/j.cobme.2019.09.009. - DOI
-
- Alvelid J, Testa I. Stable stimulated emission depletion imaging of extended sample regions. J. Phys. Appl. Phys. 2019;53:024001. doi: 10.1088/1361-6463/ab4c13. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

