Overexpression of MsRCI2D and MsRCI2E Enhances Salt Tolerance in Alfalfa (Medicago sativa L.) by Stabilizing Antioxidant Activity and Regulating Ion Homeostasis
- PMID: 36077224
- PMCID: PMC9456006
- DOI: 10.3390/ijms23179810
Overexpression of MsRCI2D and MsRCI2E Enhances Salt Tolerance in Alfalfa (Medicago sativa L.) by Stabilizing Antioxidant Activity and Regulating Ion Homeostasis
Abstract
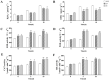
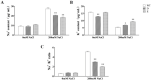
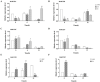
Rare cold-inducible 2 (RCI2) genes from alfalfa (Medicago sativa L.) are part of a multigene family whose members respond to a variety of abiotic stresses by regulating ion homeostasis and stabilizing membranes. In this study, salt, alkali, and ABA treatments were used to induce MsRCI2D and MsRCI2E expression in alfalfa, but the response time and the expression intensity of the MsRCI2D,-E genes were different under specific treatments. The expression intensity of the MsRCI2D gene was the highest in salt- and alkali-stressed leaves, while the MsRCI2E gene more rapidly responded to salt and ABA treatment. In addition to differences in gene expression, MsRCI2D and MsRCI2E differ in their subcellular localization. Akin to MtRCI2D from Medicago truncatula, MsRCI2D is also localized in the cell membrane, while MsRCI2E is different from MtRCI2E, localized in the cell membrane and the inner membrane. This difference might be related to an extra 20 amino acids in the C-terminal tail of MsRCI2E. We investigated the function of MsRCI2D and MsRCI2E proteins in alfalfa by generating transgenic alfalfa chimeras. Compared with the MsRCI2E-overexpressing chimera, under high-salinity stress (200 mmol·L-1 NaCl), the MsRCI2D-overexpressing chimera exhibited a better phenotype, manifested as a higher chlorophyll content and a lower MDA content. After salt treatment, the enzyme activities of SOD, POD, CAT, and GR in MsRCI2D- and -E-overexpressing roots were significantly higher than those in the control. In addition, after salt stress, the Na+ content in MsRCI2D- and -E-transformed roots was lower than that in the control; K+ was higher than that in the control; and the Na+/K+ ratio was lower than that in the control. Correspondingly, H+-ATPase, SOS1, and NHX1 genes were significantly up-regulated, and the HKT gene was significantly down-regulated after 6 h of salt treatment. MsRCI2D was also found to regulate the expression of the MsRCI2B and MsRCI2E genes, and the MsRCI2E gene could alter the expression of the MsRCI2A, MsRCI2B, and MsRCI2D genes. MsRCI2D- and -E-overexpressing alfalfa was found to have higher salt tolerance, manifested as improved activity of antioxidant enzymes, reduced content of reactive oxygen species, and sustained Na+ and K+ ion balance by regulating the expression of the H+-ATPase, SOS1, NHX1, HKT, and MsRCI2 genes.
Keywords: H+-ATPase; HKT; Medicago saliva L.; MsRCI2s; SOS1; salt tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Li C., Song T., Zhan L., Cong C., Xu H., Dong L., Cai H. Overexpression of MsRCI2A, MsRCI2B, and MsRCI2C in Alfalfa (Medicago sativa L.) Provides Different Extents of Enhanced Alkali and Salt Tolerance Due to Functional Specialization of MsRCI2s. Front. Plant Sci. 2021;12:702195. doi: 10.3389/fpls.2021.702195. - DOI - PMC - PubMed
-
- Khan I., Raza M.A., Awan S.A., Shah G.A., Rizwan M., Ali B., Tariq R., Hassan M.J., Alyemeni M.N., Brestic M. Amelioration of salt induced toxicity in pearl millet by seed priming with silver nanoparticles (AgNPs): The oxidative damage, antioxidant enzymes and ions uptake are major determinants of salt tolerant capacity. Plant Physiol. Biochem. 2020;56:221–232. doi: 10.1016/j.plaphy.2020.09.018. - DOI - PubMed
-
- Iqbal P., Ghani M.A., Ali B., Shahid M., Iqbal Q., Ziaf K., Azam M., Noor A., Cheema K.L., Ahmad J. Exogenous application of glutamic acid promotes cucumber (Cucumis sativus L.) growth under salt stress conditions. Emir. J. Food Agric. 2021;33:407–416. doi: 10.9755/ejfa.2021.v33.i5.2699. - DOI
-
- Ali B., Saleem M., Ali S., Shahid M., Sagir M., Tahir M.B., Ahmad K.Q., Jaremko M., Selim S., Hussain A., et al. Mitigation of Salinity Stress in Barley Genotypes with Variable Salt Tolerance by Application of Zinc Oxide Nanoparticles (ZnO NPs) Front. Plant Sci. 2022 doi: 10.3389/fpls.2022.973782. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

