Detailed Analysis of Dorsal-Ventral Gradients of Gene Expression in the Hippocampus of Adult Rats
- PMID: 36077346
- PMCID: PMC9456131
- DOI: 10.3390/ijms23179948
Detailed Analysis of Dorsal-Ventral Gradients of Gene Expression in the Hippocampus of Adult Rats
Abstract
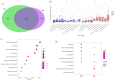
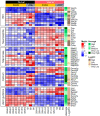
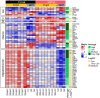
We performed RNA sequencing of the dorsal and ventral parts of the hippocampus and compared it with previously published data to determine the differences in the dorsoventral gradients of gene expression that may result from biological or technical variability. Our data suggest that the dorsal and ventral parts of the hippocampus differ in the expression of genes related to signaling pathways mediated by classical neurotransmitters (glutamate, GABA, monoamines, etc.) as well as peptide and Wnt ligands. These hippocampal parts also diverge in the expression of axon-guiding molecules (both receptors and ligands) and splice isoforms of genes associated with intercellular signaling and cell adhesion. Furthermore, analysis of differential expressions of genes specific for astrocytes, microglia, oligodendrocytes, and vascular cells suggests that non-neuronal cells may also differ in the characteristics between hippocampal parts. Analysis of expression of transposable elements showed that depletion of ribosomal RNA strongly increased the representation of transposable elements in the RNA libraries and helped to detect a weak predominance of expression of these elements in the ventral hippocampus. Our data revealed new molecular dimensions of functional differences between the dorsal and ventral hippocampus and points to possible cascades that may be involved in the longitudinal organization of the hippocampus.
Keywords: RNAseq; dorsal hippocampus; ribosomal RNA depletion; splicing; transposable elements; ventral hippocampus.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






Similar articles
-
Intracerebroventricular Administration of 192IgG-Saporin Alters Expression of Microglia-Associated Genes in the Dorsal But Not Ventral Hippocampus.Front Mol Neurosci. 2018 Jan 17;10:429. doi: 10.3389/fnmol.2017.00429. eCollection 2017. Front Mol Neurosci. 2018. PMID: 29386992 Free PMC article.
-
Differential expression of NMDA and AMPA receptor subunits in rat dorsal and ventral hippocampus.Neuroscience. 2006 Jun 19;140(1):163-75. doi: 10.1016/j.neuroscience.2006.02.003. Epub 2006 Mar 20. Neuroscience. 2006. PMID: 16542781
-
Short-term dynamics of input and output of CA1 network greatly differ between the dorsal and ventral rat hippocampus.BMC Neurosci. 2019 Jul 22;20(1):35. doi: 10.1186/s12868-019-0517-5. BMC Neurosci. 2019. PMID: 31331291 Free PMC article.
-
Hippocampal serotonin depletion facilitates the enhancement of prepulse inhibition by risperidone: possible role of 5-HT(2C) receptors in the dorsal hippocampus.Neuropharmacology. 2011 Sep;61(3):458-67. doi: 10.1016/j.neuropharm.2011.03.018. Epub 2011 Apr 5. Neuropharmacology. 2011. PMID: 21477601
-
Functional differentiation in the hippocampus.Hippocampus. 1998;8(6):608-19. doi: 10.1002/(SICI)1098-1063(1998)8:6<608::AID-HIPO3>3.0.CO;2-7. Hippocampus. 1998. PMID: 9882018 Review.
Cited by
-
Corticotropin-releasing hormone neurons control trigeminal neuralgia-induced anxiodepression via a hippocampus-to-prefrontal circuit.Sci Adv. 2024 Jan 19;10(3):eadj4196. doi: 10.1126/sciadv.adj4196. Epub 2024 Jan 19. Sci Adv. 2024. PMID: 38241377 Free PMC article.
References
-
- Nadel L. Dorsal and Ventral Hippocampal Lesions and Behavior. Physiol. Behav. 1968;3:891–900. doi: 10.1016/0031-9384(68)90174-1. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

