Identification of MADS-Box Transcription Factors in Iris laevigata and Functional Assessment of IlSEP3 and IlSVP during Flowering
- PMID: 36077350
- PMCID: PMC9456522
- DOI: 10.3390/ijms23179950
Identification of MADS-Box Transcription Factors in Iris laevigata and Functional Assessment of IlSEP3 and IlSVP during Flowering
Abstract
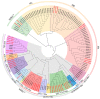
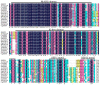
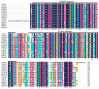
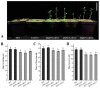
Iris laevigata is ideal for gardening and landscaping in northeast China because of its beautiful flowers and strong cold resistance. However, the short length of flowering time (2 days for individual flowers) greatly limits its applications. Molecular breeding and engineering hold high potential for producing I. laevigata of desirable flowering properties. A prerequisite is to identify and characterize key flowering control genes, the identity of which remains largely unknown in I. laevigata due to the lack of genome information. To fill this knowledge gap, we used sequencing data of the I. laevigata transcriptome to identify MADS-box gene-encoding transcription factors that have been shown to play key roles in developmental processes, including flowering. Our data revealed 41 putative MADS-box genes, which consisted of 8 type I (5 Mα and 3 Mβ, respectively) and 33 type II members (2 MIKC* and 31 MIKCC, respectively). We then selected IlSEP3 and IlSVP for functional studies and found that both are localized to the nucleus and that they interact physically in vitro. Ectopic expression of IlSEP3 in Arabidopsis resulted in early flowering (32 days) compared to that of control plants (36 days), which could be mediated by modulating the expression of FT, SOC1, AP1, SVP, SPL3, VRN1, and GA20OX. By contrast, plants overexpressing IlSVP were phenotypically similar to that of wild type. Our functional validation of IlSEP3 was consistent with the notion that SEP3 promotes flowering in multiple plant species and indicated that IlSEP3 regulates flowering in I. laevigata. Taken together, this work provided a systematic identification of MADS-box genes in I. laevigata and demonstrated that the flowering time of I. laevigata can be genetically controlled by altering the expression of key MADS-box genes.
Keywords: Arabidopsis; Iris; MADS-box; SEP3; SVP; flowering; transcription factor.
Conflict of interest statement
We declare no conflicts of interest.
Figures


Similar articles
-
Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis.Int J Mol Sci. 2021 Aug 28;22(17):9362. doi: 10.3390/ijms22179362. Int J Mol Sci. 2021. PMID: 34502271 Free PMC article.
-
Ectopic expression of LoSVP, a MADS-domain transcription factor from lily, leads to delayed flowering in transgenic Arabidopsis.Plant Cell Rep. 2020 Feb;39(2):289-298. doi: 10.1007/s00299-019-02491-1. Epub 2019 Nov 18. Plant Cell Rep. 2020. PMID: 31741036
-
Functional conservation and diversification between rice OsMADS22/OsMADS55 and Arabidopsis SVP proteins.Plant Sci. 2012 Apr;185-186:97-104. doi: 10.1016/j.plantsci.2011.09.003. Epub 2011 Sep 12. Plant Sci. 2012. PMID: 22325870
-
Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses.Plant Sci. 2011 Mar;180(3):447-53. doi: 10.1016/j.plantsci.2010.12.001. Epub 2010 Dec 14. Plant Sci. 2011. PMID: 21421391 Review.
-
The major clades of MADS-box genes and their role in the development and evolution of flowering plants.Mol Phylogenet Evol. 2003 Dec;29(3):464-89. doi: 10.1016/s1055-7903(03)00207-0. Mol Phylogenet Evol. 2003. PMID: 14615187 Review.
Cited by
-
WRKY22 Transcription Factor from Iris laevigata Regulates Flowering Time and Resistance to Salt and Drought.Plants (Basel). 2024 Apr 25;13(9):1191. doi: 10.3390/plants13091191. Plants (Basel). 2024. PMID: 38732405 Free PMC article.
-
Genome-Wide Identification of a Maize Chitinase Gene Family and the Induction of Its Expression by Fusarium verticillioides (Sacc.) Nirenberg (1976) Infection.Genes (Basel). 2024 Aug 17;15(8):1087. doi: 10.3390/genes15081087. Genes (Basel). 2024. PMID: 39202446 Free PMC article.
-
Genome-Wide Identification of MADS-Box Family Genes in Safflower (Carthamus tinctorius L.) and Functional Analysis of CtMADS24 during Flowering.Int J Mol Sci. 2023 Jan 5;24(2):1026. doi: 10.3390/ijms24021026. Int J Mol Sci. 2023. PMID: 36674539 Free PMC article.
-
Genome-wide analysis of MADS-box gene family in kiwifruit (Actinidia chinensis var. chinensis) and their potential role in floral sex differentiation.Front Genet. 2022 Nov 17;13:1043178. doi: 10.3389/fgene.2022.1043178. eCollection 2022. Front Genet. 2022. PMID: 36468015 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

