SP and KLF Transcription Factors in Cancer Metabolism
- PMID: 36077352
- PMCID: PMC9456310
- DOI: 10.3390/ijms23179956
SP and KLF Transcription Factors in Cancer Metabolism
Abstract
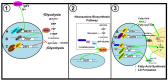
Tumor development and progression depend on reprogramming of signaling pathways that regulate cell metabolism. Alterations to various metabolic pathways such as glycolysis, oxidative phosphorylation, lipid metabolism, and hexosamine biosynthesis pathway are crucial to sustain increased redox, bioenergetic, and biosynthesis demands of a tumor cell. Transcription factors (oncogenes and tumor suppressors) play crucial roles in modulating these alterations, and their functions are tethered to major metabolic pathways under homeostatic conditions and disease initiation and advancement. Specificity proteins (SPs) and Krüppel-like factors (KLFs) are closely related transcription factors characterized by three highly conserved zinc fingers domains that interact with DNA. Studies have demonstrated that SP and KLF transcription factors are expressed in various tissues and regulate diverse processes such as proliferation, differentiation, apoptosis, inflammation, and tumorigenesis. This review highlights the role of SP and KLF transcription factors in the metabolism of various cancers and their impact on tumorigenesis. A better understanding of the role and underlying mechanisms governing the metabolic changes during tumorigenesis could provide new therapeutic opportunities for cancer treatment.
Keywords: Krüppel-like transcription factors; cancer; metabolism; specificity proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

