Identification and Characterization of Malate Dehydrogenases in Tomato (Solanum lycopersicum L.)
- PMID: 36077425
- PMCID: PMC9456053
- DOI: 10.3390/ijms231710028
Identification and Characterization of Malate Dehydrogenases in Tomato (Solanum lycopersicum L.)
Abstract
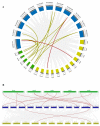
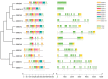
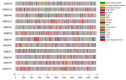
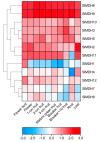
Malate dehydrogenase, which facilitates the reversible conversion of malate to oxaloacetate, is essential for energy balance, plant growth, and cold and salt tolerance. However, the genome-wide study of the MDH family has not yet been carried out in tomato (Solanum lycopersicum L.). In this study, 12 MDH genes were identified from the S. lycopersicum genome and renamed according to their chromosomal location. The tomato MDH genes were split into five groups based on phylogenetic analysis and the genes that clustered together showed similar lengths, and structures, and conserved motifs in the encoded proteins. From the 12 tomato MDH genes on the chromosomes, three pairs of segmental duplication events involving four genes were found. Each pair of genes had a Ka/Ks ratio < 1, indicating that the MDH gene family of tomato was purified during evolution. Gene expression analysis exhibited that tomato MDHs were differentially expressed in different tissues, at various stages of fruit development, and differentially regulated in response to abiotic stresses. Molecular docking of four highly expressed MDHs revealed their substrate and co-factor specificity in the reversible conversion process of malate to oxaloacetate. Further, co-localization of tomato MDH genes with quantitative trait loci (QTL) of salt stress-related phenotypes revealed their broader functions in salt stress tolerance. This study lays the foundation for functional analysis of MDH genes and genetic improvement in tomato.
Keywords: QTL mapping; abiotic stress; gene expression; genome analysis; malate dehydrogenase; molecular docking; salt stress; tomato.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

