Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas aeruginosa Model
- PMID: 36077542
- PMCID: PMC9456237
- DOI: 10.3390/ijms231710143
Liposomal Delivery of Newly Identified Prophage Lysins in a Pseudomonas aeruginosa Model
Abstract
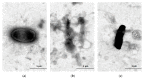
Pseudomonas aeruginosa is a Gram-negative opportunistic bacterium that presents resistance to several antibiotics, thus, representing a major threat to human and animal health. Phage-derived products, namely lysins, or peptidoglycan-hydrolyzing enzymes, can be an effective weapon against antibiotic-resistant bacteria. Whereas in Gram-positive bacteria, lysis from without is facilitated by the exposed peptidoglycan layer, this is not possible in the outer membrane-protected peptidoglycan of Gram-negative bacteria. Here, we suggest the encapsulation of lysins in liposomes as a delivery system against Gram-negative bacteria, using the model of P. aeruginosa. Bioinformatic analysis allowed for the identification of 38 distinct complete prophages within 66 P. aeruginosa genomes (16 of which newly sequenced) and led to the identification of 19 lysins of diverse sequence and function, 5 of which proceeded to wet lab analysis. The four purifiable lysins showed hydrolytic activity against Gram-positive bacterial lawns and, on zymogram assays, constituted of autoclaved P. aeruginosa cells. Additionally, lysins Pa7 and Pa119 combined with an outer membrane permeabilizer showed activity against P. aeruginosa cells. These two lysins were successfully encapsulated in DPPC:DOPE:CHEMS (molar ratio 4:4:2) liposomes with an average encapsulation efficiency of 33.33% and 32.30%, respectively. The application of the encapsulated lysins to the model P. aeruginosa led to a reduction in cell viability and resulted in cell lysis as observed in MTT cell viability assays and electron microscopy. In sum, we report here that prophages may be important sources of new enzybiotics, with prophage lysins showing high diversity and activity. In addition, these enzybiotics following their incorporation in liposomes were able to potentiate their antibacterial effect against the Gram-negative bacteria P. aeruginosa, used as the model.
Keywords: Pseudomonas aeruginosa; antibiotic resistance; bacteriophage; gram-negative bacteria; liposomes; lysins; phage therapy; prophage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Krylov V.N. Advances in Virus Research. Volume 88. Elsevier; Amsterdam, The Netherlands: 2014. Chapter Five–Bacteriophages of Pseudomonas aeruginosa: Long-Term Prospects for Use in Phage Therapy; pp. 227–278. - PubMed
-
- Ruiz-garbajosa P., Cantón R. Epidemiology of Antibiotic Resistance in Pseudomonas aeruginosa. Implications for Empiric and Definitive Therapy. Rev. Esp. Quimioter. 2017;30:8–12. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

