Anti-Tumoral Effect of Chemerin on Ovarian Cancer Cell Lines Mediated by Activation of Interferon Alpha Response
- PMID: 36077645
- PMCID: PMC9454566
- DOI: 10.3390/cancers14174108
Anti-Tumoral Effect of Chemerin on Ovarian Cancer Cell Lines Mediated by Activation of Interferon Alpha Response
Abstract
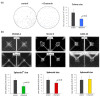
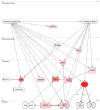
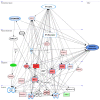
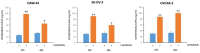
The pleiotropic adipokine chemerin affects tumor growth primarily as anti-tumoral chemoattractant inducing immunocyte recruitment. However, little is known about its effect on ovarian adenocarcinoma. In this study, we examined chemerin actions on ovarian cancer cell lines in vitro and intended to elucidate involved cell signaling mechanisms. Employing three ovarian cancer cell lines, we observed differentially pronounced effects of this adipokine. Treatment with chemerin (huChem-157) significantly reduced OVCAR-3 cell numbers (by 40.8% on day 6) and decreased the colony and spheroid growth of these cells by half. The spheroid size of SK-OV-3 ovarian cancer cells was also significantly reduced upon treatment. Transcriptome analyses of chemerin-treated cells revealed the most notably induced genes to be interferon alpha (IFNα)-response genes like IFI27, OAS1 and IFIT1 and their upstream regulator IRF9 in all cell lines tested. Finally, we found this adipokine to elevate IFNα levels about fourfold in culture medium of the employed cell lines. In conclusion, our data for the first time demonstrate IFNα as a mediator of chemerin action in vitro. The observed anti-tumoral effect of chemerin on ovarian cancer cells in vitro was mediated by the notable activation of IFNα response genes, resulting from the chemerin-triggered increase of secreted levels of this cytokine.
Keywords: adipokine; cell line; chemerin; interferon alpha; ovarian cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

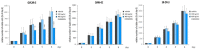





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

