A microRNA Prognostic Signature in Patients with Diffuse Intrinsic Pontine Gliomas through Non-Invasive Liquid Biopsy
- PMID: 36077842
- PMCID: PMC9454461
- DOI: 10.3390/cancers14174307
A microRNA Prognostic Signature in Patients with Diffuse Intrinsic Pontine Gliomas through Non-Invasive Liquid Biopsy
Abstract
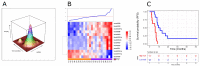
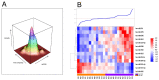
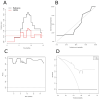
Diffuse midline gliomas (DMGs) originate in the thalamus, brainstem, cerebellum and spine. This entity includes tumors that infiltrate the pons, called diffuse intrinsic pontine gliomas (DIPGs), with a rapid onset and devastating neurological symptoms. Since surgical removal in DIPGs is not feasible, the purpose of this study was to profile circulating miRNA expression in DIPG patients in an effort to identify a non-invasive prognostic signature with clinical impact. Using a high-throughput platform, miRNA expression was profiled in serum samples collected at the time of MRI diagnosis and prior to radiation and/or systemic therapy from 47 patients enrolled in clinical studies, combining nimotuzumab and vinorelbine with concomitant radiation. With progression-free survival as the primary endpoint, a semi-supervised learning approach was used to identify a signature that was also tested taking overall survival as the clinical endpoint. A signature comprising 13 circulating miRNAs was identified in the training set (n = 23) as being able to stratify patients by risk of disease progression (log-rank p = 0.00014; HR = 7.99, 95% CI 2.38-26.87). When challenged in a separate validation set (n = 24), it confirmed its ability to predict progression (log-rank p = 0.00026; HR = 5.51, 95% CI 2.03-14.9). The value of our signature was also confirmed when overall survival was considered (log-rank p = 0.0021, HR = 4.12, 95% CI 1.57-10.8). We have identified and validated a prognostic marker based on the expression of 13 circulating miRNAs that can shed light on a patient's risk of progression. This is the first demonstration of the usefulness of nucleic acids circulating in the blood as powerful, easy-to-assay molecular markers of disease status in DIPG. This study provides Class II evidence that a signature based on 13 circulating miRNAs is associated with the risk of disease progression.
Keywords: circulating miRNAs; diffuse intrinsic pontine gliomas; neuro-oncology; prognosis.
Conflict of interest statement
The authors have no conflicts of interest to disclose. The funders had no role in the design of the study, nor in the collection, analysis and interpretation of the data, in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. - DOI - PMC - PubMed
-
- Beuriat P.A., Szathmari A., Di Rocco F., Kanold J., Mottolese C., Frappaz D. Diffuse intrinsic pontine glioma in children: Document or treat? World Neurosurg. 2016;93:485.e11–485.e14. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

