Identification and Characterization of Alternative Splicing Variants and Positive Selection Genes Related to Distinct Growth Rates of Antlers Using Comparative Transcriptome Sequencing
- PMID: 36077923
- PMCID: PMC9454627
- DOI: 10.3390/ani12172203
Identification and Characterization of Alternative Splicing Variants and Positive Selection Genes Related to Distinct Growth Rates of Antlers Using Comparative Transcriptome Sequencing
Abstract
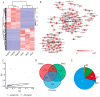
The molecular mechanism underlying rapid antler growth has not been elucidated. The contrast of the wapiti and sika deer antler provides a potential model for comparative studies for the identification of potent growth factors and unique regulatory systems. In the present study, reference transcriptomes of antler RM tissue of wapiti and sika deer were constructed using single molecule real time sequencing data. The expression profiling, positive selection, and alternative splicing of the antler transcripts were compared. The results showed that: a total of 44,485 reference full-length transcripts of antlers were obtained; 254 highly expressed transcripts (HETs) and 1936 differentially expressed genes (DEGs) were enriched and correlated principally with translation, endochondral ossification and ribosome; 228 genes were found to be under strong positive selection and would thus be important for the evolution of wapiti and sika deer; among the alternative splicing variants, 381 genes were annotated; and 4 genes with node degree values greater than 50 were identified through interaction network analysis. We identified a negative and a positive regulator for rapid antler growth, namely RNA Binding Motif Protein X-Linked (RBMX) and methyltransferase-like 3 (METTL3), respectively. Overall, we took advantage of this significant difference in growth rate and performed the comparative analyses of the antlers to identify key specific factors that might be candidates for the positive or negative regulation of phenomenal antler growth rate.
Keywords: alternative splicing variants; antler growth rate; comparative transcriptome; positive selection genes; single molecule real time sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Comparative transcriptome analysis of the main beam and brow tine of sika deer antler provides insights into the molecular control of rapid antler growth.Cell Mol Biol Lett. 2020 Sep 7;25:42. doi: 10.1186/s11658-020-00234-9. eCollection 2020. Cell Mol Biol Lett. 2020. PMID: 32944020 Free PMC article.
-
Full-length transcriptome and microRNA sequencing reveal the specific gene-regulation network of velvet antler in sika deer with extremely different velvet antler weight.Mol Genet Genomics. 2019 Apr;294(2):431-443. doi: 10.1007/s00438-018-1520-8. Epub 2018 Dec 11. Mol Genet Genomics. 2019. PMID: 30539301
-
Comparative analysis of differentially expressed genes in Sika deer antler at different stages.Mol Biol Rep. 2013 Feb;40(2):1665-76. doi: 10.1007/s11033-012-2216-5. Epub 2012 Oct 18. Mol Biol Rep. 2013. PMID: 23073784
-
Deer antlers: a zoological curiosity or the key to understanding organ regeneration in mammals?J Anat. 2005 Nov;207(5):603-18. doi: 10.1111/j.1469-7580.2005.00478.x. J Anat. 2005. PMID: 16313394 Free PMC article. Review.
-
Exploring the mechanisms regulating regeneration of deer antlers.Philos Trans R Soc Lond B Biol Sci. 2004 May 29;359(1445):809-22. doi: 10.1098/rstb.2004.1471. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15293809 Free PMC article. Review.
References
-
- Goss R.J. Deer Antlers: Regeneration, Function and Evolution. Academic Press; New York, NY, USA: 1983.
-
- Bartos L., Bahbouh R. Antler size and fluctuating asymmetry in red deer (Cervus elaphus) stags and probability of becoming a harem holder in rut. Biol. J. Linn. Soc. 2006;87:59–68. doi: 10.1111/j.1095-8312.2006.00555.x. - DOI
-
- Liu W.J. Remarks on Chinese materia medica—Young Pilose antler—A precious drug. Zhong Yao Tong Bao. 1981;6:43–45. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

