Monoamine Oxidase-Dependent Pro-Survival Signaling in Diabetic Hearts Is Mediated by miRNAs
- PMID: 36078109
- PMCID: PMC9454570
- DOI: 10.3390/cells11172697
Monoamine Oxidase-Dependent Pro-Survival Signaling in Diabetic Hearts Is Mediated by miRNAs
Abstract
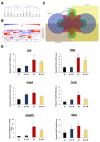
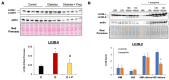
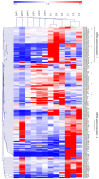
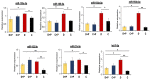
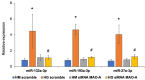
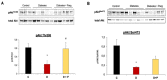
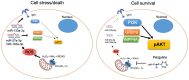
Diabetes leads to cardiomyopathy and heart failure, the leading cause of death for diabetic patients. Monoamine oxidase (MAO) inhibition in diabetic cardiomyopathy prevents oxidative stress, mitochondrial and endoplasmic reticulum stress and the development of diastolic dysfunction. However, it is unclear whether, in addition to the direct effects exerted on the mitochondria, MAO activity is able to post-transcriptionally regulate cardiomyocyte function and survival in diabetes. To this aim, we performed gene and miRNA expression profiling in cardiac tissue from streptozotocin-treated mice (model of type 1 diabetes (T1D)), administered with either vehicle or MAOs inhibitor pargyline for 12 weeks. We found that inhibition of MAO activity in T1D hearts leads to profound transcriptomic changes, affecting autophagy and pro-survival pathways activation. MAO activity in T1D hearts increased miR-133a-3p, -193a-3p and -27a-3p expression. These miRNAs target insulin-like growth factor receptor 1 (Igf1r), growth factor receptor bound protein 10 and inositol polyphosphate 4 phosphatase type 1A, respectively, all components of the IGF1R/PI3K/AKT signaling pathway. Indeed, AKT activation was significantly downregulated in T1D hearts, whereas MAO inhibition restored the activation of this pro-survival pathway. The present study provides an important link between MAO activity, transcriptomic changes and activation of pro-survival signaling and autophagy in diabetic cardiomyopathy.
Keywords: autophagy; diabetic cardiomyopathy; miRNAs; monoamine oxidase; pro-survival pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rota M., Le Capitaine N., Hosoda T., Boni A., De Angelis A., Padin-Iruegas M.E., Esposito G., Vitale S., Urbanek K., Casarsa C., et al. Diabetes promotes cardiac stem cell aging and heart failure, which are prevented by deletion of the p66shc gene. Circ. Res. 2006;99:42–52. doi: 10.1161/01.RES.0000231289.63468.08. - DOI - PubMed
-
- Giorgio M., Migliaccio E., Orsini F., Paolucci D., Moroni M., Contursi C., Pelliccia G., Luzi L., Minucci S., Marcaccio M., et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. - DOI - PubMed
-
- Carpi A., Menabo R., Kaludercic N., Pelicci P., Di Lisa F., Giorgio M. The cardioprotective effects elicited by p66(Shc) ablation demonstrate the crucial role of mitochondrial ROS formation in ischemia/reperfusion injury. Biochim. Biophys. Acta. 2009;1787:774–780. doi: 10.1016/j.bbabio.2009.04.001. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

