Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L
- PMID: 36079595
- PMCID: PMC9460571
- DOI: 10.3390/plants11172213
Molecular Cloning and Expression Analysis of the Cryptochrome Gene CiPlant-CRY1 in Antarctic Ice Alga Chlamydomonas sp. ICE-L
Abstract
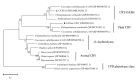
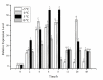
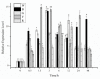
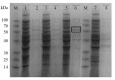
Cryptochrome (CRY) is a kind of flavin-binding protein that can sense blue light and near-ultraviolet light, and participates in the light response of organisms and the regulation of the circadian clock. The complete open reading frame (ORF) of CiPlant-CRY1 (GenBank ID OM389130.1), encoding one kind of CRY, was cloned from the Antarctic ice alga Chlamydomonas sp. ICE-L. The quantitative real-time PCR study showed that the expression level of the CiPlant-CRY1 gene was the highest at 5 °C and salinity of 32‱. CiPlant-CRY1 was positively regulated by blue or yellow light, suggesting that it is involved in the establishment of photomorphology. The CiPlant-CRY1 gene can respond to polar day and polar night, indicating its expression is regulated by circadian rhythm. The expression level of CiPlant-CRY1 was most affected by UVB irradiation, which may be related to the adaptation of ice algae to a strong ultraviolet radiation environment. Moreover, the recombinant protein of CiPlant-CRY1 was expressed by prokaryotic expression. This study may be important for exploring the light-induced rhythm regulation of Antarctic ice algae in the polar marine environment.
Keywords: Chlamydomonas sp. ICE-L; CiPlant-CRY1; cryptochrome; extreme Antarctic environments; heterologous expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources

