Phylogenomics and Systematics of Overlooked Mesoamerican and South American Polyploid Broad-Leaved Festuca Grasses Differentiate F. sects. Glabricarpae and Ruprechtia and F. subgen. Asperifolia, Erosiflorae, Mallopetalon and Coironhuecu (subgen. nov.)
- PMID: 36079685
- PMCID: PMC9460391
- DOI: 10.3390/plants11172303
Phylogenomics and Systematics of Overlooked Mesoamerican and South American Polyploid Broad-Leaved Festuca Grasses Differentiate F. sects. Glabricarpae and Ruprechtia and F. subgen. Asperifolia, Erosiflorae, Mallopetalon and Coironhuecu (subgen. nov.)
Abstract
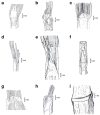
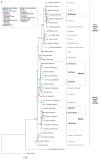
Allopolyploidy is considered a driver of diversity in subtribe Loliinae. We investigate the evolution and systematics of the poorly studied Mesoamerican and South American polyploid broad-leaved Festuca L. species of uncertain origin and unclear taxonomy. A taxonomic study of seven diagnostic morphological traits was conducted on a representation of 22 species. Phylogenomic analyses were performed on a representation of these supraspecific taxa and all other Loliinae lineages using separate data from the entire plastome, nuclear rDNA 45S and 5S genes, and repetitive DNA elements. F. subgen. Mallopetalon falls within the fine-leaved (FL) Loliinae clade, whereas the remaining taxa are nested within the broad-leaved (BL) Loliinae clade forming two separate Mexico-Central-South American (MCSAI, MCSAII) lineages. MCSAI includes representatives of F. sect. Glabricarpae and F. subgen. Asperifolia plus F. superba, and MCSAII of F. subgen. Erosiflorae and F. sect. Ruprechtia plus F. argentina. MCSAII likely had a BL Leucopoa paternal ancestor, MCSAI and MCSAII a BL Meso-South American maternal ancestor, and Mallopetalon FL, American I-II ancestors. Plastome vs. nuclear topological discordances corroborated the hybrid allopolyploid origins of these taxa, some of which probably originated from Northern Hemisphere ancestors. The observed data indicate rapid reticulate radiations in the Central-South American subcontinent. Our systematic study supports the reclassification of some studied taxa in different supraspecific Festuca ranks.
Keywords: Mexico–Central–South American broad-leaved Festuca; allopolyploid speciation; phylogeny; plastome; rDNA 45S and 5S genes; repeatome; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

