Deciphering the Potential of Pre and Pro-Vitamin D of Mushrooms against Mpro and PLpro Proteases of COVID-19: An In Silico Approach
- PMID: 36080385
- PMCID: PMC9458008
- DOI: 10.3390/molecules27175620
Deciphering the Potential of Pre and Pro-Vitamin D of Mushrooms against Mpro and PLpro Proteases of COVID-19: An In Silico Approach
Abstract
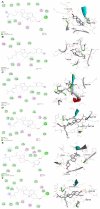
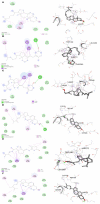
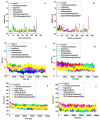
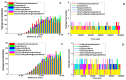
Vitamin D's role in combating the SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), the virus causing COVID-19, has been established in unveiling viable inhibitors of COVID-19. The current study investigated the role of pre and pro-vitamin D bioactives from edible mushrooms against Mpro and PLpro proteases of SARS-CoV-2 by computational experiments. The bioactives of mushrooms, specifically ergosterol (provitamin D2), 7-dehydrocholesterol (provitamin-D3), 22,23-dihydroergocalciferol (provitamin-D4), cholecalciferol (vitamin-D3), and ergocalciferol (vitamin D2) were screened against Mpro and PLpro. Molecular docking analyses of the generated bioactive protease complexes unravelled the differential docking energies, which ranged from -7.5 kcal/mol to -4.5 kcal/mol. Ergosterol exhibited the lowest binding energy (-7.5 kcal/mol) against Mpro and PLpro (-5.9 kcal/mol). The Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) and MD simulation analyses indicated that the generated complexes were stable, thus affirming the putative binding of the bioactives to viral proteases. Considering the pivotal role of vitamin D bioactives, their direct interactions against SARS-CoV-2 proteases highlight the promising role of bioactives present in mushrooms as potent nutraceuticals against COVID-19.
Keywords: SARS-CoV-2; edible mushrooms; in-silico studies; pre-vitamin-D; pro-vitamin-D.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

