Identification, biogenesis, function, and mechanism of action of circular RNAs in plants
- PMID: 36081344
- PMCID: PMC9860190
- DOI: 10.1016/j.xplc.2022.100430
Identification, biogenesis, function, and mechanism of action of circular RNAs in plants
Abstract
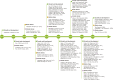
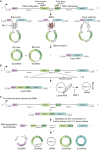
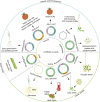
Circular RNAs (circRNAs) are a class of single-stranded, closed RNA molecules with unique functions that are ubiquitously expressed in all eukaryotes. The biogenesis of circRNAs is regulated by specific cis-acting elements and trans-acting factors in humans and animals. circRNAs mainly exert their biological functions by acting as microRNA sponges, forming R-loops, interacting with RNA-binding proteins, or being translated into polypeptides or proteins in human and animal cells. Genome-wide identification of circRNAs has been performed in multiple plant species, and the results suggest that circRNAs are abundant and ubiquitously expressed in plants. There is emerging compelling evidence to suggest that circRNAs play essential roles during plant growth and development as well as in the responses to biotic and abiotic stress. However, compared with recent advances in human and animal systems, the roles of most circRNAs in plants are unclear at present. Here we review the identification, biogenesis, function, and mechanism of action of plant circRNAs, which will provide a fundamental understanding of the characteristics and complexity of circRNAs in plants.
Keywords: biogenesis; function; identification; mechanism; plant circRNA.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Aktaş T., Avşar Ilık İ., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. - PubMed
-
- Bai J.f., Guo H.y., Yuan S.h., Li T.t., Duan W.j., Liu Z.h., Li Y.m., Zhang T.b., Zhang F.t., Liao X.z., et al. Uncovering ceRNA integrated networks that associate with fertility in a photoperiod and temperature sensitive male sterile wheat line. Biotechnol. Biotechnol. Equip. 2021;35:1317–1330.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

