Characterization of MET Exon 14 Skipping Alterations (in NSCLC) and Identification of Potential Therapeutic Targets Using Whole Transcriptome Sequencing
- PMID: 36082279
- PMCID: PMC9445394
- DOI: 10.1016/j.jtocrr.2022.100381
Characterization of MET Exon 14 Skipping Alterations (in NSCLC) and Identification of Potential Therapeutic Targets Using Whole Transcriptome Sequencing
Abstract
Introduction: Genomic alterations in the juxtamembrane exon 14 splice sites in NSCLC lead to increased MET stability and oncogenesis. We present the largest cohort study of MET Exon 14 (METex14) using whole transcriptome sequencing.
Methods: A total of 21,582 NSCLC tumor samples underwent complete genomic profiling with next-generation sequencing of DNA (592 Gene Panel, NextSeq, whole exome sequencing, NovaSeq) and RNA (NovaSeq, whole transcriptome sequencing). Clinicopathologic information including programmed death-ligand 1 and tumor mutational burden were collected and RNA expression for mutation subtypes and MET amplification were quantified. Immunogenic signatures and potential pathways of invasion were characterized using single-sample gene set enrichment analysis and mRNA gene signatures.
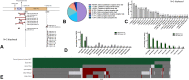
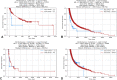
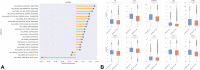
Results: A total of 533tumors (2.47%) with METex14 were identified. The most common alterations were point mutations (49.5%) at donor splice sites. Most alterations translated to increased MET expression, with MET co-amplification resulting in synergistic increase in expression (q < 0.05). Common coalterations were amplifications of MDM2 (19.0% versus 1.8% wild-type [WT]), HMGA2 (13.2% versus 0.98% WT), and CDK4 (10.0% versus 1.5% WT) (q < 0.05). High programmed death-ligand 1 > 50% (52.5% versus 27.3% WT, q < 0.0001) and lower proportion of high tumor mutational burden (>10 mutations per megabase, 8.3% versus 36.7% WT, p < 0.0001) were associated with METex14, which were also enriched in both immunogenic signatures and immunosuppressive checkpoints. Pathways associated with METex14 included angiogenesis and apical junction pathways (q < 0.05).
Conclusions: METex14 splicing alterations and MET co-amplification translated to higher and synergistic MET expression at the transcriptomic level. High frequencies of MDM2 and CDK4 co-amplifications and association with multiple immunosuppressive checkpoints and angiogenic pathways provide insight into potential actionable targets for combination strategies in METex14 NSCLC.
Keywords: Immune signatures; MDM2; METex14; Non–small cell lung cancer; RNA expression; Whole transcriptome sequencing.
© 2022 The Authors.
Figures


References
-
- Ma P.C., Jagadeeswaran R., Jagadeesh S., et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–1488. - PubMed
-
- Frampton G.M., Ali S.M., Rosenzweig M., et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. - PubMed
-
- Schrock A.B., Frampton G.M., Suh J., et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol. 2016;11:1493–1502. - PubMed
-
- Awad M.M., Oxnard G.R., Jackman D.M., et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-met overexpression. J Clin Oncol. 2016;34:721–730. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

