The bacterial effector GarD shields Chlamydia trachomatis inclusions from RNF213-mediated ubiquitylation and destruction
- PMID: 36084633
- PMCID: PMC9772000
- DOI: 10.1016/j.chom.2022.08.008
The bacterial effector GarD shields Chlamydia trachomatis inclusions from RNF213-mediated ubiquitylation and destruction
Abstract
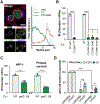
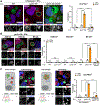
Chlamydia trachomatis is the leading cause of sexually transmitted bacterial infections and a major threat to women's reproductive health in particular. This obligate intracellular pathogen resides and replicates within a cellular compartment termed an inclusion, where it is sheltered by unknown mechanisms from gamma-interferon (IFNγ)-induced cell-autonomous host immunity. Through a genetic screen, we uncovered the Chlamydia inclusion membrane protein gamma resistance determinant (GarD) as a bacterial factor protecting inclusions from cell-autonomous immunity. In IFNγ-primed human cells, inclusions formed by garD loss-of-function mutants become decorated with linear ubiquitin and are eliminated. Leveraging cellular genome-wide association data, we identified the ubiquitin E3 ligase RNF213 as a candidate anti-Chlamydia protein. We demonstrate that IFNγ-inducible RNF213 facilitates the ubiquitylation and destruction of GarD-deficient inclusions. Furthermore, we show that GarD operates as a cis-acting stealth factor barring RNF213 from targeting inclusions, thus functionally defining GarD as an RNF213 antagonist essential for chlamydial growth during IFNγ-stimulated immunity.
Keywords: Chlamydia; NDP52; RNF213; TAX1BP1; autophagy; cell-autonomous immunity; interferon; interferon-stimulated genes; optineurin; ubiquitylation.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Aiyar A, Quayle AJ, Buckner LR, Sherchand SP, Chang TL, Zea AH, Martin DH, and Belland RJ (2014). Influence of the tryptophan-indole-IFNgamma axis on human genital Chlamydia trachomatis infection: role of vaginal co-infections. Front Cell Infect Microbiol 4, 72. 10.3389/fcimb.2014.00072. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

