Mutations in SCNM1 cause orofaciodigital syndrome due to minor intron splicing defects affecting primary cilia
- PMID: 36084634
- PMCID: PMC9606384
- DOI: 10.1016/j.ajhg.2022.08.009
Mutations in SCNM1 cause orofaciodigital syndrome due to minor intron splicing defects affecting primary cilia
Abstract
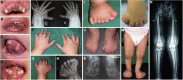
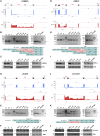
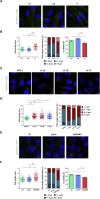
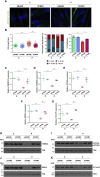
Orofaciodigital syndrome (OFD) is a genetically heterogeneous ciliopathy characterized by anomalies of the oral cavity, face, and digits. We describe individuals with OFD from three unrelated families having bi-allelic loss-of-function variants in SCNM1 as the cause of their condition. SCNM1 encodes a protein recently shown to be a component of the human minor spliceosome. However, so far the effect of loss of SCNM1 function on human cells had not been assessed. Using a comparative transcriptome analysis between fibroblasts derived from an OFD-affected individual harboring SCNM1 mutations and control fibroblasts, we identified a set of genes with defective minor intron (U12) processing in the fibroblasts of the affected subject. These results were reproduced in SCNM1 knockout hTERT RPE-1 (RPE-1) cells engineered by CRISPR-Cas9-mediated editing and in SCNM1 siRNA-treated RPE-1 cultures. Notably, expression of TMEM107 and FAM92A encoding primary cilia and basal body proteins, respectively, and that of DERL2, ZC3H8, and C17orf75, were severely reduced in SCNM1-deficient cells. Primary fibroblasts containing SCNM1 mutations, as well as SCNM1 knockout and SCNM1 knockdown RPE-1 cells, were also found with abnormally elongated cilia. Conversely, cilia length and expression of SCNM1-regulated genes were restored in SCNM1-deficient fibroblasts following reintroduction of SCNM1 via retroviral delivery. Additionally, functional analysis in SCNM1-retrotransduced fibroblasts showed that SCNM1 is a positive mediator of Hedgehog (Hh) signaling. Our findings demonstrate that defective U12 intron splicing can lead to a typical ciliopathy such as OFD and reveal that primary cilia length and Hh signaling are regulated by the minor spliceosome through SCNM1 activity.
Keywords: SCNM1; U12 introns; ciliopathy; hedgehog signaling; minor spliceosome; orofaciodigital syndrome; primary cilia.
Copyright © 2022 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

