Leveraging genomic diversity for discovery in an electronic health record linked biobank: the UCLA ATLAS Community Health Initiative
- PMID: 36085083
- PMCID: PMC9461263
- DOI: 10.1186/s13073-022-01106-x
Leveraging genomic diversity for discovery in an electronic health record linked biobank: the UCLA ATLAS Community Health Initiative
Erratum in
-
Author Correction: Leveraging genomic diversity for discovery in an electronic health record linked biobank: the UCLA ATLAS Community Health Initiative.Genome Med. 2022 Nov 16;14(1):128. doi: 10.1186/s13073-022-01128-5. Genome Med. 2022. PMID: 36384576 Free PMC article. No abstract available.
Abstract
Background: Large medical centers in urban areas, like Los Angeles, care for a diverse patient population and offer the potential to study the interplay between genetic ancestry and social determinants of health. Here, we explore the implications of genetic ancestry within the University of California, Los Angeles (UCLA) ATLAS Community Health Initiative-an ancestrally diverse biobank of genomic data linked with de-identified electronic health records (EHRs) of UCLA Health patients (N=36,736).
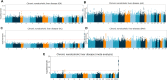
Methods: We quantify the extensive continental and subcontinental genetic diversity within the ATLAS data through principal component analysis, identity-by-descent, and genetic admixture. We assess the relationship between genetically inferred ancestry (GIA) and >1500 EHR-derived phenotypes (phecodes). Finally, we demonstrate the utility of genetic data linked with EHR to perform ancestry-specific and multi-ancestry genome and phenome-wide scans across a broad set of disease phenotypes.
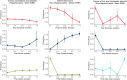
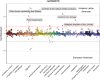
Results: We identify 5 continental-scale GIA clusters including European American (EA), African American (AA), Hispanic Latino American (HL), South Asian American (SAA) and East Asian American (EAA) individuals and 7 subcontinental GIA clusters within the EAA GIA corresponding to Chinese American, Vietnamese American, and Japanese American individuals. Although we broadly find that self-identified race/ethnicity (SIRE) is highly correlated with GIA, we still observe marked differences between the two, emphasizing that the populations defined by these two criteria are not analogous. We find a total of 259 significant associations between continental GIA and phecodes even after accounting for individuals' SIRE, demonstrating that for some phenotypes, GIA provides information not already captured by SIRE. GWAS identifies significant associations for liver disease in the 22q13.31 locus across the HL and EAA GIA groups (HL p-value=2.32×10-16, EAA p-value=6.73×10-11). A subsequent PheWAS at the top SNP reveals significant associations with neurologic and neoplastic phenotypes specifically within the HL GIA group.
Conclusions: Overall, our results explore the interplay between SIRE and GIA within a disease context and underscore the utility of studying the genomes of diverse individuals through biobank-scale genotyping linked with EHR-based phenotyping.
Keywords: Biobank; Electronic health records; Genetic ancestry; Genome-wide association studies; Phenome-wide association studies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures