Inhibition of the Niemann-Pick C1 protein is a conserved feature of multiple strains of pathogenic mycobacteria
- PMID: 36085278
- PMCID: PMC9463166
- DOI: 10.1038/s41467-022-32553-0
Inhibition of the Niemann-Pick C1 protein is a conserved feature of multiple strains of pathogenic mycobacteria
Abstract
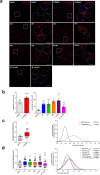
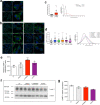
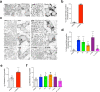
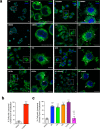
Mycobacterium tuberculosis (Mtb) survives and replicates within host macrophages (MΦ) and subverts multiple antimicrobial defense mechanisms. Previously, we reported that lipids shed by pathogenic mycobacteria inhibit NPC1, the lysosomal membrane protein deficient in the lysosomal storage disorder Niemann-Pick disease type C (NPC). Inhibition of NPC1 leads to a drop in lysosomal calcium levels, blocking phagosome-lysosome fusion leading to mycobacterial survival. We speculated that the production of specific cell wall lipid(s) that inhibit NPC1 could have been a critical step in the evolution of pathogenicity. We therefore investigated whether lipid extracts from clinical Mtb strains from multiple Mtb lineages, Mtb complex (MTBC) members and non-tubercular mycobacteria (NTM) inhibit the NPC pathway. We report that inhibition of the NPC pathway was present in all clinical isolates from Mtb lineages 1, 2, 3 and 4, Mycobacterium bovis and the NTM, Mycobacterium abscessus and Mycobacterium avium. However, lipid extract from Mycobacterium canettii, which is considered to resemble the common ancestor of the MTBC did not inhibit the NPC1 pathway. We conclude that the evolution of NPC1 inhibitory mycobacterial cell wall lipids evolved early and post divergence from Mycobacterium canettii-related mycobacteria and that this activity contributes significantly to the promotion of disease.
© 2022. The Author(s).
Conflict of interest statement
F.M.P. is a consultant to and co-founder of IntraBio. The other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

