The lysosomal proteome of senescent cells contributes to the senescence secretome
- PMID: 36087066
- PMCID: PMC9577959
- DOI: 10.1111/acel.13707
The lysosomal proteome of senescent cells contributes to the senescence secretome
Abstract
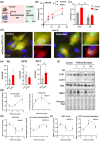
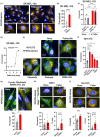
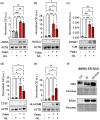
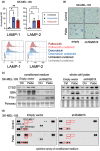
Senescent cells accumulate in tissues over time, favoring the onset and progression of multiple age-related diseases. Senescent cells present a remarkable increase in lysosomal mass and elevated autophagic activity. Here, we report that two main autophagic pathways macroautophagy (MA) and chaperone-mediated autophagy (CMA) are constitutively upregulated in senescent cells. Proteomic analyses of the subpopulations of lysosomes preferentially engaged in each of these types of autophagy revealed profound quantitative and qualitative changes in senescent cells, affecting both lysosomal resident proteins and cargo proteins delivered to lysosomes for degradation. These studies have led us to identify resident lysosomal proteins that are highly augmented in senescent cells and can be used as novel markers of senescence, such as arylsulfatase ARSA. The abundant secretome of senescent cells, known as SASP, is considered their main pathological mediator; however, little is known about the mechanisms of SASP secretion. Some secretory cells, including melanocytes, use the small GTPase RAB27A to perform lysosomal secretion. We found that this process is exacerbated in the case of senescent melanoma cells, as revealed by the exposure of lysosomal membrane integral proteins LAMP1 and LAMP2 in their plasma membrane. Interestingly, a subset of SASP components, including cytokines CCL2, CCL3, CXCL12, cathepsin CTSD, or the protease inhibitor SERPINE1, are secreted in a RAB27A-dependent manner in senescent melanoma cells. Finally, proteins previously identified as plasma biomarkers of aging are highly enriched in the lysosomes of senescent cells, including CTSD. We conclude that the lysosomal proteome of senescent cells is profoundly reconfigured, and that some senescent cells can be highly active in lysosomal exocytosis.
Keywords: SASP; aging; autophagy; cellular senescence; exocytosis; lysosome.
© 2022 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
A.M.C. is co‐founder of Selphagy Therapeutics (now under LifeBiosciences, Boston, MA, USA) and consults for Generian Therapeutics and Cognition. M.S. is shareholder and advisor of Senolytic Therapeutics, Inc., Rejuveron Senescence Therapeutics, AG, and Altos Labs, Inc; and is shareholder of Life Biosciences, Inc. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Figures



References
-
- Auteri, J. S. , Okada, A. , Bochaki, V. , & Fred Dice, J. (1983). Regulation of intracellular protein degradation in IMR90 human diploid fibroblasts. Journal of Cellular Physiology, 115, 167–174. - PubMed
-
- Blott, E. J. , & Griffiths, G. M. (2002). Secretory lysosomes. Nature Reviews. Molecular Cell Biology, 3, 122–131. - PubMed
-
- Bourdenx, M. , Martín‐Segura, A. , Scrivo, A. , Rodriguez‐Navarro, J. A. , Kaushik, S. , Tasset, I. , Diaz, A. , Storm, N. J. , Xin, Q. , Juste, Y. R. , Stevenson, E. , Luengo, E. , Clement, C. C. , Choi, S. J. , Krogan, N. J. , Mosharov, E. V. , Santambrogio, L. , Grueninger, F. , Collin, L. , … Cuervo, A. M. (2021). Chaperone‐mediated autophagy prevents collapse of the neuronal metastable proteome. Cell, 184, 2696–2714.e25. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

