Functional genomics uncovers the transcription factor BNC2 as required for myofibroblastic activation in fibrosis
- PMID: 36088459
- PMCID: PMC9464213
- DOI: 10.1038/s41467-022-33063-9
Functional genomics uncovers the transcription factor BNC2 as required for myofibroblastic activation in fibrosis
Abstract
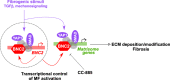
Tissue injury triggers activation of mesenchymal lineage cells into wound-repairing myofibroblasts, whose unrestrained activity leads to fibrosis. Although this process is largely controlled at the transcriptional level, whether the main transcription factors involved have all been identified has remained elusive. Here, we report multi-omics analyses unraveling Basonuclin 2 (BNC2) as a myofibroblast identity transcription factor. Using liver fibrosis as a model for in-depth investigations, we first show that BNC2 expression is induced in both mouse and human fibrotic livers from different etiologies and decreases upon human liver fibrosis regression. Importantly, we found that BNC2 transcriptional induction is a specific feature of myofibroblastic activation in fibrotic tissues. Mechanistically, BNC2 expression and activities allow to integrate pro-fibrotic stimuli, including TGFβ and Hippo/YAP1 signaling, towards induction of matrisome genes such as those encoding type I collagen. As a consequence, Bnc2 deficiency blunts collagen deposition in livers of mice fed a fibrogenic diet. Additionally, our work establishes BNC2 as potentially druggable since we identified the thalidomide derivative CC-885 as a BNC2 inhibitor. Altogether, we propose that BNC2 is a transcription factor involved in canonical pathways driving myofibroblastic activation in fibrosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

