Prokaryotes of renowned Karlovy Vary (Carlsbad) thermal springs: phylogenetic and cultivation analysis
- PMID: 36089611
- PMCID: PMC9465906
- DOI: 10.1186/s40793-022-00440-2
Prokaryotes of renowned Karlovy Vary (Carlsbad) thermal springs: phylogenetic and cultivation analysis
Abstract
Background: The extreme conditions of thermal springs constitute a unique aquatic habitat characterized by low nutrient contents and the absence of human impacts on the microbial community composition. Thus, these springs may host phylogenetically novel microorganisms with potential use in biotechnology. With this hypothesis in mind, we examined the microbial composition of four thermal springs of the world-renowned spa town of Karlovy Vary (Carlsbad), Czechia, which differ in their temperature and chemical composition.
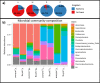
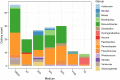
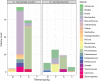
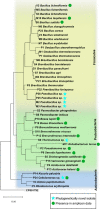
Results: Microbial profiling using 16S rRNA gene sequencing revealed the presence of phylogenetically novel taxa at various taxonomic levels, spanning from genera to phyla. Many sequences belonged to novel classes within the phyla Hydrothermae, Altiarchaeota, Verrucomicrobia, and TA06. Cultivation-based methods employing oligotrophic media resulted in the isolation of 44 unique bacterial isolates. These include strains that withstand concentrations of up to 12% NaClw/v in cultivation media or survive a temperature of 100 °C, as well as hitherto uncultured bacterial species belonging to the genera Thermomonas, Paenibacillus, and Cellulomonas. These isolates harbored stress response genes that allow them to thrive in the extreme environment of thermal springs.
Conclusions: Our study is the first to analyze the overall microbial community composition of the renowned Karlovy Vary thermal springs. We provide insight into yet another level of uniqueness of these springs. In addition to their unique health benefits and cultural significance, we demonstrate that these springs harbor phylogenetically distinct microorganisms with unusual life strategies. Our findings open up avenues for future research with the promise of a deeper understanding of the metabolic potential of these microorganisms.
Keywords: Amplicon sequencing analysis; Cultivation analysis; Phylogenetic novelty; Thermal water springs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Culturomics of Bacteria from Radon-Saturated Water of the World's Oldest Radium Mine.Microbiol Spectr. 2022 Oct 26;10(5):e0199522. doi: 10.1128/spectrum.01995-22. Epub 2022 Aug 24. Microbiol Spectr. 2022. PMID: 36000901 Free PMC article.
-
Untapped bacterial diversity and metabolic potential within Unkeshwar hot springs, India.Arch Microbiol. 2018 Jul;200(5):753-770. doi: 10.1007/s00203-018-1484-4. Epub 2018 Feb 2. Arch Microbiol. 2018. PMID: 29396619
-
Diversity and Activity of Sulfate-Reducing Prokaryotes in Kamchatka Hot Springs.Microorganisms. 2021 Oct 1;9(10):2072. doi: 10.3390/microorganisms9102072. Microorganisms. 2021. PMID: 34683394 Free PMC article.
-
Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review.Microorganisms. 2021 Jul 9;9(7):1473. doi: 10.3390/microorganisms9071473. Microorganisms. 2021. PMID: 34361908 Free PMC article. Review.
-
Bacterial diversity in 110 thermal hot springs of Indian Himalayan Region (IHR).3 Biotech. 2022 Sep;12(9):238. doi: 10.1007/s13205-022-03270-8. Epub 2022 Aug 21. 3 Biotech. 2022. PMID: 36003895 Free PMC article. Review.
Cited by
-
Ecological versatility and biotechnological promise: Comprehensive characterization of the isolated thermophilic Bacillus strains.PLoS One. 2024 Apr 18;19(4):e0297217. doi: 10.1371/journal.pone.0297217. eCollection 2024. PLoS One. 2024. PMID: 38635692 Free PMC article.
-
Compost, plants and endophytes versus metal contamination: choice of a restoration strategy steers the microbiome in polymetallic mine waste.Environ Microbiome. 2023 Oct 7;18(1):74. doi: 10.1186/s40793-023-00528-3. Environ Microbiome. 2023. PMID: 37805609 Free PMC article.
-
Reaching unreachables: Obstacles and successes of microbial cultivation and their reasons.Front Microbiol. 2023 Mar 7;14:1089630. doi: 10.3389/fmicb.2023.1089630. eCollection 2023. Front Microbiol. 2023. PMID: 36960281 Free PMC article. Review.
-
Characterization of Archaea membrane lipids in radioactive springs using shotgun lipidomics.Folia Microbiol (Praha). 2025 Feb;70(1):225-233. doi: 10.1007/s12223-024-01235-3. Epub 2024 Dec 17. Folia Microbiol (Praha). 2025. PMID: 39688758
-
The dynamic history of prokaryotic phyla: discovery, diversity and division.Int J Syst Evol Microbiol. 2024 Sep;74(9):006508. doi: 10.1099/ijsem.0.006508. Int J Syst Evol Microbiol. 2024. PMID: 39250184 Review.
References
-
- Alfaro C, Wallace M. Origin and classification of springs and historical review with current applications. Environ Geol. 1994;24(2):112–124. doi: 10.1007/BF00767884. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
