Development of a multi-recombinase polymerase amplification assay for rapid identification of COVID-19, influenza A and B
- PMID: 36089764
- PMCID: PMC9538624
- DOI: 10.1002/jmv.28139
Development of a multi-recombinase polymerase amplification assay for rapid identification of COVID-19, influenza A and B
Abstract
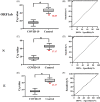
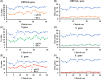
The coronavirus disease 2019 (COVID-19) pandemic caused extensive loss of life worldwide. Further, the COVID-19 and influenza mix-infection had caused great distress to the diagnosis of the disease. To control illness progression and limit viral spread within the population, a real-time reverse-transcription PCR (RT-PCR) assay for early diagnosis of COVID-19 was developed, but detection was time-consuming (4-6 h). To improve the diagnosis of COVID-19 and influenza, we herein developed a recombinase polymerase amplification (RPA) method for simple and rapid amplification of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19 and Influenza A (H1N1, H3N2) and B (influenza B). Genes encoding the matrix protein (M) for H1N1, and the hemagglutinin (HA) for H3N2, and the polymerase A (PA) for Influenza B, and the nucleocapsid protein (N), the RNA-dependent-RNA polymerase (RdRP) in the open reading frame 1ab (ORF1ab) region, and the envelope protein (E) for SARS-CoV-2 were selected, and specific primers were designed. We validated our method using SARS-CoV-2, H1N1, H3N2 and influenza B plasmid standards and RNA samples extracted from COVID-19 and Influenza A/B (RT-PCR-verified) positive patients. The method could detect SARS-CoV-2 plasmid standard DNA quantitatively between 102 and 105 copies/ml with a log linearity of 0.99 in 22 min. And this method also be very effective in simultaneous detection of H1N1, H3N2 and influenza B. Clinical validation of 100 cases revealed a sensitivity of 100% for differentiating COVID-19 patients from healthy controls when the specificity was set at 90%. These results demonstrate that this nucleic acid testing method is advantageous compared with traditional PCR and other isothermal nucleic acid amplification methods in terms of time and portability. This method could potentially be used for detection of SARS-CoV-2, H1N1, H3N2 and influenza B, and adapted for point-of-care (POC) detection of a broad range of infectious pathogens in resource-limited settings.
Keywords: COVID-19; RPA; SARS-CoV-2; influenza; laboratory diagnosis.
© 2022 The Authors. Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

