NIfTI-MRS: A standard data format for magnetic resonance spectroscopy
- PMID: 36089825
- PMCID: PMC7613677
- DOI: 10.1002/mrm.29418
NIfTI-MRS: A standard data format for magnetic resonance spectroscopy
Abstract
Purpose: Multiple data formats in the MRS community currently hinder data sharing and integration. NIfTI-MRS is proposed as a standard spectroscopy data format, implemented as an extension to the Neuroimaging informatics technology initiative (NIfTI) format. This standardized format can facilitate data sharing and algorithm development as well as ease integration of MRS analysis alongside other imaging modalities.
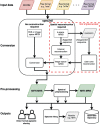
Methods: A file format using the NIfTI header extension framework incorporates essential spectroscopic metadata and additional encoding dimensions. A detailed description of the specification is provided. An open-source command-line conversion program is implemented to convert single-voxel and spectroscopic imaging data to NIfTI-MRS. Visualization of data in NIfTI-MRS is provided by development of a dedicated plugin for FSLeyes, the FMRIB Software Library (FSL) image viewer.
Results: Online documentation and 10 example datasets in the proposed format are provided. Code examples of NIfTI-MRS readers are implemented in common programming languages. Conversion software, spec2nii, currently converts 14 formats where data is stored in image-space to NIfTI-MRS, including Digital Imaging and Communications in Medicine (DICOM) and vendor proprietary formats.
Conclusion: NIfTI-MRS aims to solve issues arising from multiple data formats being used in the MRS community. Through a single conversion point, processing and analysis of MRS data are simplified, thereby lowering the barrier to use of MRS. Furthermore, it can serve as the basis for open data sharing, collaboration, and interoperability of analysis programs. Greater standardization and harmonization become possible. By aligning with the dominant format in neuroimaging, NIfTI-MRS enables the use of mature tools present in the imaging community, demonstrated in this work by using a dedicated imaging tool, FSLeyes, for visualization.
Keywords: MRS; MRSI; open data format; spectroscopy; visualization.
© 2022 The Authors. Magnetic Resonance in Medicine published by Wiley Periodicals LLC on behalf of International Society for Magnetic Resonance in Medicine.
Figures




References
-
- Palombo M, Shemesh N, Ronen I, Valette J. Insights into brain microstructure from in vivo DW‐MRS. Neuroimage. 2018;182:97‐116. - PubMed
-
- C.8.14 MR Spectroscopy Modules. Available at: http://dicom.nema.org/medical/dicom/2017c/output/chtml/part03/sect_C.8.1.... Accessed December 16, 2020.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

