Comparison of extraction methods for intracellular metabolomics of human tissues
- PMID: 36090025
- PMCID: PMC9461704
- DOI: 10.3389/fmolb.2022.932261
Comparison of extraction methods for intracellular metabolomics of human tissues
Abstract
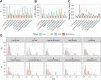
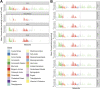
Analyses of metabolic compounds inside cells or tissues provide high information content since they represent the endpoint of biological information flow and are a snapshot of the integration of many regulatory processes. However, quantification of the abundance of metabolites requires their careful extraction. We present a comprehensive study comparing ten extraction protocols in four human sample types (liver tissue, bone marrow, HL60, and HEK cells) aiming to detect and quantify up to 630 metabolites of different chemical classes. We show that the extraction efficiency and repeatability are highly variable across protocols, tissues, and chemical classes of metabolites. We used different quality metrics including the limit of detection and variability between replicates as well as the sum of concentrations as a global estimate of analytical repeatability of the extraction. The coverage of extracted metabolites depends on the used solvents, which has implications for the design of measurements of different sample types and metabolic compounds of interest. The benchmark dataset can be explored in an easy-to-use, interactive, and flexible online resource (R/shiny app MetaboExtract: http://www.metaboextract.shiny.dkfz.de) for context-specific selection of the optimal extraction method. Furthermore, data processing and conversion functionality underlying the shiny app are accessible as an R package: https://cran.r-project.org/package=MetAlyzer.
Keywords: absolute quantification; extraction protocol; intra-cellular; metabolism; metabolomics.
Copyright © 2022 Andresen, Boch, Gegner, Mechtel, Narr, Birgin, Rasbach, Rahbari, Trumpp, Poschet and Hübschmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Banerjee S., Zare R. N., Tibshirani R. J., Kunder C. A., Nolley R., Fan R., et al. (2017). Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. U. S. A. 114, 3334–3339. 10.1073/pnas.1700677114 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

