Framework From a Multidisciplinary Approach for Transitioning Variants of Unknown Significance From Clinical Genetic Testing in Kidney Disease to a Definitive Classification
- PMID: 36090499
- PMCID: PMC9459028
- DOI: 10.1016/j.ekir.2022.06.014
Framework From a Multidisciplinary Approach for Transitioning Variants of Unknown Significance From Clinical Genetic Testing in Kidney Disease to a Definitive Classification
Abstract
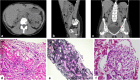
Introduction: Monogenic causes in over 300 kidney-associated genes account for approximately 12% of end stage kidney disease (ESKD) cases. Advances in sequencing and large customized panels enable the noninvasive diagnosis of monogenic kidney disease at relatively low cost, thereby allowing for more precise management for patients and their families. A major challenge is interpreting rare variants, many of which are classified as variants of unknown significance (VUS). We present a framework in which we thoroughly evaluated and provided evidence of pathogenicity for HNF1B-p.Arg303His, a VUS returned from clinical diagnostic testing for a kidney transplant candidate.
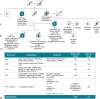
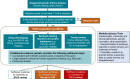
Methods: A blueprint was designed by a multidisciplinary team of clinicians, molecular biologists, and diagnostic geneticists. The blueprint included using a health system-based cohort with genetic and clinical information to perform deep phenotyping of VUS heterozygotes to identify the candidate VUS and rule out other VUS, examination of existing genetic databases, as well as functional testing.
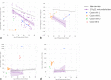
Results: Our approach demonstrated evidence for pathogenicity for HNF1B-p.Arg303His by showing similar burden of kidney manifestations in this variant to known HNF1B pathogenic variants, and greater burden compared to noncarriers.
Conclusion: Determination of a molecular diagnosis for the example family allows for proper surveillance and management of HNF1B-related manifestations such as kidney disease, diabetes, and hypomagnesemia with important implications for safe living-related kidney donation. The candidate gene-variant pair also allows for clinical biomarker testing for aberrations of linked pathways. This working model may be applicable to other diseases of genetic etiology.
Keywords: HNF1B-MODY; autosomal dominant tubulointerstitial kidney disease (ADTKD); chronic kidney disease; genetics; hypomagnesemia; pancreatitis.
© 2022 International Society of Nephrology. Published by Elsevier Inc.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

