Extending and using anatomical vocabularies in the stimulating peripheral activity to relieve conditions project
- PMID: 36090663
- PMCID: PMC9449460
- DOI: 10.3389/fninf.2022.819198
Extending and using anatomical vocabularies in the stimulating peripheral activity to relieve conditions project
Abstract
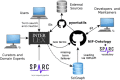
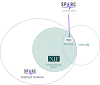
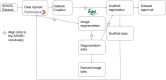
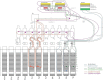
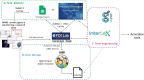
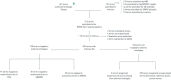
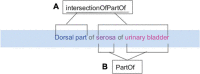
The stimulating peripheral activity to relieve conditions (SPARC) program is a US National Institutes of Health-funded effort to improve our understanding of the neural circuitry of the autonomic nervous system (ANS) in support of bioelectronic medicine. As part of this effort, the SPARC project is generating multi-species, multimodal data, models, simulations, and anatomical maps supported by a comprehensive knowledge base of autonomic circuitry. To facilitate the organization of and integration across multi-faceted SPARC data and models, SPARC is implementing the findable, accessible, interoperable, and reusable (FAIR) data principles to ensure that all SPARC products are findable, accessible, interoperable, and reusable. We are therefore annotating and describing all products with a common FAIR vocabulary. The SPARC Vocabulary is built from a set of community ontologies covering major domains relevant to SPARC, including anatomy, physiology, experimental techniques, and molecules. The SPARC Vocabulary is incorporated into tools researchers use to segment and annotate their data, facilitating the application of these ontologies for annotation of research data. However, since investigators perform deep annotations on experimental data, not all terms and relationships are available in community ontologies. We therefore implemented a term management and vocabulary extension pipeline where SPARC researchers may extend the SPARC Vocabulary using InterLex, an online vocabulary management system. To ensure the quality of contributed terms, we have set up a curated term request and review pipeline specifically for anatomical terms involving expert review. Accepted terms are added to the SPARC Vocabulary and, when appropriate, contributed back to community ontologies to enhance ANS coverage. Here, we provide an overview of the SPARC Vocabulary, the infrastructure and process for implementing the term management and review pipeline. In an analysis of >300 anatomical contributed terms, the majority represented composite terms that necessitated combining terms within and across existing ontologies. Although these terms are not good candidates for community ontologies, they can be linked to structures contained within these ontologies. We conclude that the term request pipeline serves as a useful adjunct to community ontologies for annotating experimental data and increases the FAIRness of SPARC data.
Keywords: InterLex; SPARC; anatomical terms; ontologies; peripheral nervous system (PNS).
Copyright © 2022 Surles-Zeigler, Sincomb, Gillespie, de Bono, Bresnahan, Mawe, Grethe, Tappan, Heal and Martone.
Conflict of interest statement
MM and JG have an equity interest in SciCrunch, Inc., a company that may potentially benefit from the research results. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. ST and MH were employed by MBF Bioscience, the creator of software referenced in this manuscript. BB was employed by the company Whitby et al., Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Angstman P. J., Tappan S. J., Sullivan A. E., Thomas G. C., Rodriguez A., Hoppes D. M., et al. (2020). Neuromorphological File Specification. Williston: MBF Bioscience.
-
- Bandrowski A., Grethe J. S., Pilko A., Gillespie T., Pine G., Patel B., et al. (2021). SPARC Data Structure: Rationale and Design of a FAIR Standard for Biomedical Research Data. bioRxiv. [Preprint]. 10.1101/2021.02.10.430563 - DOI
-
- Bartholomew D. (2012). Mariadb vs. mysql. Dostopano 7:2014.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

