Lineage BA.2 dominated the Omicron SARS-CoV-2 epidemic wave in the Philippines
- PMID: 36090771
- PMCID: PMC9452094
- DOI: 10.1093/ve/veac078
Lineage BA.2 dominated the Omicron SARS-CoV-2 epidemic wave in the Philippines
Abstract
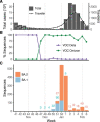
The Omicron severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant led to a dramatic global epidemic wave following detection in South Africa in November 2021. The BA.1 Omicron lineage was dominant and responsible for most SARS-CoV-2 outbreaks in countries around the world during December 2021-January 2022, while other Omicron lineages, including BA.2, accounted for the minority of global isolates. Here, we describe the Omicron wave in the Philippines by analysing genomic data. Our results identify the presence of both BA.1 and BA.2 lineages in the Philippines in December 2021, before cases surged in January 2022. We infer that only the BA.2 lineage underwent sustained transmission in the country, with an estimated emergence around 18 November 2021 (95 per cent highest posterior density: 6-28 November), while despite multiple introductions, BA.1 transmission remained limited. These results suggest that the Philippines was one of the earliest areas affected by BA.2 and reiterate the importance of whole genome sequencing for monitoring outbreaks.
Keywords: Philippines; SARS-CoV-2; genomic epidemiology.
© The Author(s) 2022. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


References
-
- Aksamentov I. et al. (2021) ‘Nextclade: Clade Assignment, Mutation Calling and Quality Control for Viral Genomes’, Journal of Open Source Software, 6: 3773.
-
- Department of Health (Department of Health), Philippines . COVID-19 Tracker. <https://doh.gov.ph/covid19tracker> accessed 25 Feb 2022.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

