Molecular characterization and functional analysis of DIGIRR from golden pompano (Trachinotus ovatus)
- PMID: 36091048
- PMCID: PMC9448908
- DOI: 10.3389/fimmu.2022.974310
Molecular characterization and functional analysis of DIGIRR from golden pompano (Trachinotus ovatus)
Abstract
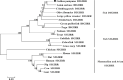
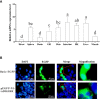
Mammalian single immunoglobulin (Ig) interleukin-1 receptor related molecule (SIGIRR), an important member of the Toll/interleukin-1 receptor (TIR) family, plays important balancing roles in the inflammatory responses. In the present study, the double Ig interleukin-1 receptor related molecule (DIGIRR), the homologous of SIGIRR, was characterized in golden pompano (Trachinotus ovatus) (termed as trDIGIRR). The full-length cDNA of trDIGIRR was 2,167 bp with an open reading frame (ORF) of 1,572 bp encoding 523 amino acids. The trDIGIRR contained several conserved domains including a signal peptide, two Ig domains, a transmembrane domain and a TIR domain, and shared high sequence identities with its teleost counterparts. Realtime qPCR analysis revealed that the trDIGIRR was distributed in all tissues examined, with high expressions in intestine, liver and head kidney. The expressions of trDIGIRR were induced by Vibrio alginolyticus, lipopolysaccharide (LPS) and polyinosinic-polycytidylic acid (poly I:C) challenge. Further analysis revealed that trDIGIRR was mainly located in the cytoplasm. In addition, the co-immunoprecipitation (co-IP) assay identified that trDIGIRR could interact with myeloid differentiation factor 88 (MyD88), but not interact with TIR domain containing adaptor protein inducing interferon-β (TRIF). Our results provide basis for studying the immune role of fish DIGIRR.
Keywords: DIGIRR; Trachinotus ovatus; co-IP; immune response; subcellular localization.
Copyright © 2022 Xie, Gao, Cao, Ji, Zhang, Wei and Qi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Bell E. Innate immunity-TLR signalling. Nat Rev Immunol (2003) 3(9):692–2. doi: 10.1038/nri1185 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

