Precise somatic genome editing for treatment of inborn errors of immunity
- PMID: 36091069
- PMCID: PMC9459235
- DOI: 10.3389/fimmu.2022.960348
Precise somatic genome editing for treatment of inborn errors of immunity
Abstract
Rapid advances in high throughput sequencing have substantially expedited the identification and diagnosis of inborn errors of immunity (IEI). Correction of faulty genes in the hematopoietic stem cells can potentially provide cures for the majority of these monogenic immune disorders. Given the clinical efficacies of vector-based gene therapies already established for certain groups of IEI, the recently emerged genome editing technologies promise to bring safer and more versatile treatment options. Here, we review the latest development in genome editing technologies, focusing on the state-of-the-art tools with improved precision and safety profiles. We subsequently summarize the recent preclinical applications of genome editing tools in IEI models, and discuss the major challenges and future perspectives of such treatment modalities. Continued explorations of precise genome editing for IEI treatment shall move us closer toward curing these unfortunate rare diseases.
Keywords: base editing; gene therapy; genome editing; hematopoietic stem cell (HSC); inborn errors of immunity (IEI); prime editing.
Copyright © 2022 Meng, Sun and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
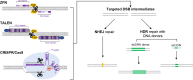
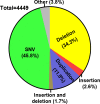


References
-
- Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, et al. Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J Clin Immunol (2020) 40(1):24–64. doi: 10.1007/s10875-019-00737-x - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

