Exosomes Derived hsa-miR-4669 as a Novel Biomarker for Early Predicting the Response of Subcutaneous Immunotherapy in Pediatric Allergic Rhinitis
- PMID: 36091336
- PMCID: PMC9451037
- DOI: 10.2147/JIR.S379414
Exosomes Derived hsa-miR-4669 as a Novel Biomarker for Early Predicting the Response of Subcutaneous Immunotherapy in Pediatric Allergic Rhinitis
Abstract
Purpose: Subcutaneous immunotherapy (SCIT) is an effective treatment for pediatric allergic rhinitis (AR), but its efficacy fluctuates among individuals. This study aims to identify the profile of serum exosomes derived microRNAs (miRNAs) and evaluate their capacities to early predict SCIT efficacy in pediatric AR.
Patients and methods: High-throughput sequencing was applied to identify the miRNA of serum exosomes in AR children. GO enrichment and KEGG pathway analysis were performed to enrich the biological annotations of target mRNAs of miRNAs. Then we validated differentially expressed miRNAs in two independent cohorts by RT-qPCR. Logistic regression and receiver operating characteristic curve (ROC) were applied to evaluate the abilities of identified miRNAs in predicting the efficacy of SCIT in AR children.
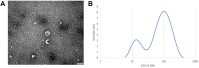
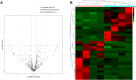
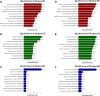
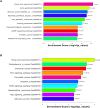
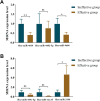
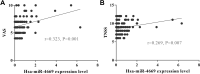
Results: A total of 812 miRNAs were detected in the serum exosomes, including 16 upregulated and 14 downregulated. Differentially expressed genes are enriched in the biological process of developmental process and regulation of cellular process, and gathered in pathways such as the signaling pathways regulating pluripotency of stem cells and the Wnt signaling pathway. In the first validation cohort, hsa-miR-4669 (P=0.009) and hsa-miR-4686 (P=0.032) were significantly downregulated in the effective group than the ineffective group, while hsa-miR-3196 (P=0.015) was upregulated. In the second cohort, hsa-miR-4669 level (P<0.0001) was downregulated in the effective group than the ineffective group. In addition, logistic regression revealed that hsa-miR-4669 level was correlated with the visual analogue scale (r=0.323, P=0.001) and total nasal symptoms score (r=0.269, P =0.007). ROC curve highlighted that hsa-miR-4669 level exhibited a reliable accuracy in predicting SCIT efficacy in pediatric AR (AUC=0.785).
Conclusion: Serum exosomes derived miRNA were associated with the efficacy of SCIT. Serum exosomes derived hsa-miR-4669 might serve as a novel biomarker for early predicting the response of SCIT in AR children.
Keywords: allergic rhinitis; children; exosome; miRNA; subcutaneous immunotherapy.
© 2022 Jiang et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures

Similar articles
-
Serum exosomal miR-146a-3p associates with disease severity and efficacy of sublingual immunotherapy in allergic rhinitis.Int Immunopharmacol. 2023 Mar;116:109777. doi: 10.1016/j.intimp.2023.109777. Epub 2023 Jan 28. Int Immunopharmacol. 2023. PMID: 36716518
-
Identification of Robust Biomarkers for Early Predicting Efficacy of Subcutaneous Immunotherapy in Children With House Dust Mite-Induced Allergic Rhinitis by Multiple Cytokine Profiling.Front Immunol. 2022 Jan 12;12:805404. doi: 10.3389/fimmu.2021.805404. eCollection 2021. Front Immunol. 2022. PMID: 35095890 Free PMC article.
-
Circulating exosomal has-miR-24-3p and has-miR-128-3p reflect early efficacy of sublingual immunotherapy in allergic rhinitis.Int Immunopharmacol. 2023 Nov;124(Pt A):110822. doi: 10.1016/j.intimp.2023.110822. Epub 2023 Aug 22. Int Immunopharmacol. 2023. PMID: 37619414
-
Circulating C-X-C Motif Ligand 13 as a Biomarker for Early Predicting Efficacy of Subcutaneous Immunotherapy in Children With Chronic Allergic Rhinitis.Front Pediatr. 2022 May 4;10:872152. doi: 10.3389/fped.2022.872152. eCollection 2022. Front Pediatr. 2022. PMID: 35601415 Free PMC article.
-
Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes.Clin Epigenetics. 2017 Dec 12;9:127. doi: 10.1186/s13148-017-0428-1. eCollection 2017. Clin Epigenetics. 2017. PMID: 29255496 Free PMC article.
Cited by
-
Overexpression of miR-4669 Enhances Tumor Aggressiveness and Generates an Immunosuppressive Tumor Microenvironment in Hepatocellular Carcinoma: Its Clinical Value as a Predictive Biomarker.Int J Mol Sci. 2023 Apr 26;24(9):7908. doi: 10.3390/ijms24097908. Int J Mol Sci. 2023. PMID: 37175615 Free PMC article.
-
Endometrial Mesenchymal Stem Cell-Derived Exosomal miR-4669 Promotes EMT in Adenomyosis by Inducing M2 Macrophage Polarization via the DUSP6/ERK Pathway.Reprod Sci. 2025 Aug 6. doi: 10.1007/s43032-025-01944-1. Online ahead of print. Reprod Sci. 2025. PMID: 40770601
-
A review of recent advances in exosomes and allergic rhinitis.Front Pharmacol. 2022 Dec 15;13:1096984. doi: 10.3389/fphar.2022.1096984. eCollection 2022. Front Pharmacol. 2022. PMID: 36588711 Free PMC article. Review.
-
Circulating Exosomal microRNA Profiles Associated with Risk of Postoperative Recurrence in Chronic Rhinosinusitis with Nasal Polyps.J Inflamm Res. 2024 Aug 23;17:5619-5631. doi: 10.2147/JIR.S472963. eCollection 2024. J Inflamm Res. 2024. PMID: 39193125 Free PMC article.
-
Application of Extracellular Vesicles in Allergic Rhinitis: A Systematic Review.Int J Mol Sci. 2022 Dec 26;24(1):367. doi: 10.3390/ijms24010367. Int J Mol Sci. 2022. PMID: 36613810 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

