LncRNA-mRNA modules involved in goat rumen development: Insights from genome-wide transcriptome profiling
- PMID: 36091364
- PMCID: PMC9449361
- DOI: 10.3389/fphys.2022.979121
LncRNA-mRNA modules involved in goat rumen development: Insights from genome-wide transcriptome profiling
Abstract
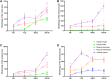
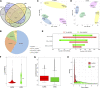
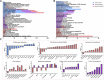
The rumen is an essential digestive and absorption organ of ruminants. During fetal life, lactation, and post-weaning period, goat rumen undergoes drastic morphological and metabolic-functional changes triggered by potential regulated genes and non-coding RNA molecules. As the essential regulatory factors, long non-coding RNAs (lncRNAs) have vital functions in various biological activities. However, their roles during rumen development are still poorly explored in ruminants. To explore the genome-wide expression profiles of lncRNAs and mRNAs in the goat rumens, we generated 5,007 lncRNAs and 19,738 mRNAs identified during the fetal and prepubertal stages by the high-throughput RNA sequencing. Notably, 365 lncRNAs and 2,877 mRNAs were considered to be differentially expressed. The weighted gene co-expression network analysis and functional analysis were performed to explore the regulatory roles of those differentially expressed molecules. The cis-and trans-target genes of differently expressed lncRNAs were enriched for pathways related to focal adhesion, cGMP-PKG signaling pathway, alpha-linolenic acid metabolism, arachidonic acid metabolism, and fat digestion and absorption. Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes analyses showed that the differently expressed genes mainly participated in mitotic cytokinesis, desmosome, fatty acid degradation, cell adhesion molecules, and fatty acid metabolism. The prediction of lncRNA-mRNA interaction networks further revealed transcripts potentially involved in rumen development. The present study profiles a global overview of lncRNAs and mRNAs during rumen development. Our findings provide valuable resources for genetic regulation and molecular mechanisms of rumen development in ruminants.
Keywords: Goat; different expression; lncRNA; mRNA; pathway; rumen; transcriptome.
Copyright © 2022 Zhong, Zhao, Zhan, Wang, Cao, Dai, Guo, Li, Zhang and Niu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Bondzio A., Gabler C., Badewien-Rentzsch B., Schulze P., Martens H., Einspanier R. (2011). Identification of differentially expressed proteins in ruminal epithelium in response to a concentrate-supplemented diet. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G260–G268. 10.1152/ajpgi.00304.2010 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

