A species-specific lncRNA modulates the reproductive ability of the asian tiger mosquito
- PMID: 36091452
- PMCID: PMC9448860
- DOI: 10.3389/fbioe.2022.885767
A species-specific lncRNA modulates the reproductive ability of the asian tiger mosquito
Abstract
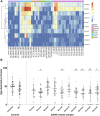
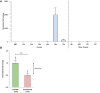
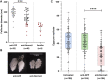
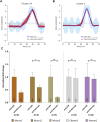
Long non-coding RNA (lncRNA) research has emerged as an independent scientific field in recent years. Despite their association with critical cellular and metabolic processes in plenty of organisms, lncRNAs are still a largely unexplored area in mosquito research. We propose that they could serve as exceptional tools for pest management due to unique features they possess. These include low inter-species sequence conservation and high tissue specificity. In the present study, we investigated the role of ovary-specific lncRNAs in the reproductive ability of the Asian tiger mosquito, Aedes albopictus. Through the analysis of transcriptomic data, we identified several lncRNAs that were differentially expressed upon blood feeding; we called these genes Norma (NOn-coding RNA in Mosquito ovAries). We observed that silencing some of these Normas resulted in significant impact on mosquito fecundity and fertility. We further focused on Norma3 whose silencing resulted in 43% oviposition reduction, in smaller ovaries and 53% hatching reduction of the laid eggs, compared to anti-GFP controls. Moreover, a significant downregulation of 2 mucins withing a neighboring (∼100 Kb) mucin cluster was observed in smaller anti-Norma3 ovaries, indicating a potential mechanism of in-cis regulation between Norma3 and the mucins. Our work constitutes the first experimental proof-of-evidence connecting lncRNAs with mosquito reproduction and opens a novel path for pest management.
Keywords: Aedes albopictus; RNAi pest control; lncRNAs (long non-coding RNAs); species-specific control; tiger mosquito.
Copyright © 2022 Belavilas-Trovas, Gregoriou, Tastsoglou, Soukia, Giakountis and Mathiopoulos.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genome-wide identification of Aedes albopictus long noncoding RNAs and their association with dengue and Zika virus infection.PLoS Negl Trop Dis. 2021 Jan 22;15(1):e0008351. doi: 10.1371/journal.pntd.0008351. eCollection 2021 Jan. PLoS Negl Trop Dis. 2021. PMID: 33481791 Free PMC article.
-
The Insect Growth Regulator Pyriproxyfen Terminates Egg Diapause in the Asian Tiger Mosquito, Aedes albopictus.PLoS One. 2015 Jun 19;10(6):e0130499. doi: 10.1371/journal.pone.0130499. eCollection 2015. PLoS One. 2015. PMID: 26090954 Free PMC article.
-
Differentiation of Long Non-Coding RNA and mRNA Expression Profiles in Male and Female Aedes albopictus.Front Genet. 2019 Oct 14;10:975. doi: 10.3389/fgene.2019.00975. eCollection 2019. Front Genet. 2019. PMID: 31681418 Free PMC article.
-
The Eye of the Tiger, the Thrill of the Fight: Effective Larval and Adult Control Measures Against the Asian Tiger Mosquito, Aedes albopictus (Diptera: Culicidae), in North America.J Med Entomol. 2016 Sep;53(5):1029-47. doi: 10.1093/jme/tjw096. Epub 2016 Jun 28. J Med Entomol. 2016. PMID: 27354440 Review.
-
Practical management plan for invasive mosquito species in Europe: I. Asian tiger mosquito (Aedes albopictus).Travel Med Infect Dis. 2020 May-Jun;35:101691. doi: 10.1016/j.tmaid.2020.101691. Epub 2020 Apr 22. Travel Med Infect Dis. 2020. PMID: 32334085 Review.
Cited by
-
Transcriptome reveals the roles and potential mechanisms of CeRNA in the regulation of salivary gland development in the tick Rhipicephalus haemaphysaloides.Front Cell Infect Microbiol. 2025 Apr 30;15:1573239. doi: 10.3389/fcimb.2025.1573239. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40370407 Free PMC article.
-
A comparison of normalization methods for the expression of genes associated with oxidative stress in the liver of sheep.BMC Genom Data. 2025 Jul 31;26(1):53. doi: 10.1186/s12863-025-01345-y. BMC Genom Data. 2025. PMID: 40745265 Free PMC article.
-
A deeper insight into the sialome of male and female Culex quinquefasciatus mosquitoes.BMC Genomics. 2023 Mar 20;24(1):135. doi: 10.1186/s12864-023-09236-1. BMC Genomics. 2023. PMID: 36941562 Free PMC article.
-
Ovarian transcriptome analyses indicate that weak juvenile hormone signaling underlies the molecular basis of oogenesis deficiencies in mosquitoes.BMC Biol. 2025 Jun 9;23(1):160. doi: 10.1186/s12915-025-02266-z. BMC Biol. 2025. PMID: 40484957 Free PMC article.
-
A longitudinal transcriptomic analysis from unfed to post-engorgement midguts of adult female Ixodes scapularis.Sci Rep. 2023 Jul 13;13(1):11360. doi: 10.1038/s41598-023-38207-5. Sci Rep. 2023. PMID: 37443274 Free PMC article. Review.
References
-
- Angelini P., Macini P., Finarelli A., Po C., Venturelli C., Bellini R., et al. (2008). Chikungunya epidemic outbreak in Emilia-Romagna (Italy) during summer 2007. Parassitologia 50 (1/2), 97–98. - PubMed
LinkOut - more resources
Full Text Sources

