Evaluation of pathways to the C-glycosyl isoflavone puerarin in roots of kudzu (Pueraria montana lobata)
- PMID: 36091880
- PMCID: PMC9438399
- DOI: 10.1002/pld3.442
Evaluation of pathways to the C-glycosyl isoflavone puerarin in roots of kudzu (Pueraria montana lobata)
Abstract
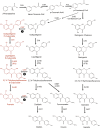
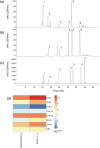
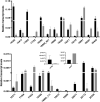
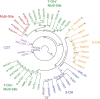
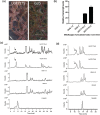
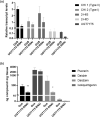
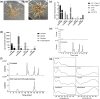
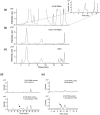
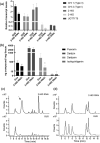
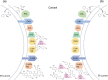
Kudzu (Pueraria montana lobata) is used as a traditional medicine in China and Southeast Asia but is a noxious weed in the Southeastern United States. It produces both O- and C-glycosylated isoflavones, with puerarin (C-glucosyl daidzein) as an important bioactive compound. Currently, the stage of the isoflavone pathway at which the C-glycosyl unit is added remains unclear, with a recent report of direct C-glycosylation of daidzein contradicting earlier labeling studies supporting C-glycosylation at the level of chalcone. We have employed comparative mRNA sequencing of the roots from two Pueraria species, one of which produces puerarin (field collected P. montana lobata) and one of which does not (commercial Pueraria phaseoloides), to identify candidate uridine diphosphate glycosyltransferase (UGT) enzymes involved in puerarin biosynthesis. Expression of recombinant UGTs in Escherichia coli and candidate C-glycosyltransferases in Medicago truncatula were used to explore substrate specificities, and gene silencing of UGT and key isoflavone biosynthetic genes in kudzu hairy roots employed to test hypotheses concerning the substrate(s) for C-glycosylation. Our results confirm UGT71T5 as a C-glycosyltransferase of isoflavone biosynthesis in kudzu. Enzymatic, isotope labeling, and genetic analyses suggest that puerarin arises both from the direct action of UGT71T5 on daidzein and via a second route in which the C-glycosidic linkage is introduced to the chalcone isoliquiritigenin.
© 2022 The Authors. Plant Direct published by American Society of Plant Biologists and the Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The Authors did not report any conflict of interest.
Figures

References
-
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
LinkOut - more resources
Full Text Sources

