Screening for potential undiagnosed Gaucher disease patients: Utilisation of the Gaucher earlier diagnosis consensus point-scoring system (GED-C PSS) in conjunction with electronic health record data, tissue specimens, and small nucleotide polymorphism (SNP) genotype data available in Finnish biobanks
- PMID: 36092251
- PMCID: PMC9449642
- DOI: 10.1016/j.ymgmr.2022.100911
Screening for potential undiagnosed Gaucher disease patients: Utilisation of the Gaucher earlier diagnosis consensus point-scoring system (GED-C PSS) in conjunction with electronic health record data, tissue specimens, and small nucleotide polymorphism (SNP) genotype data available in Finnish biobanks
Abstract
Background: Autosomal recessive Gaucher disease (GD) is likely underdiagnosed in many countries. Because the number of diagnosed GD patients in Finland is relatively low, and the true prevalence is currently not known, it was hypothesized that undiagnosed GD patients may exist in Finland. Our previous study demonstrated the applicability of Gaucher Earlier Diagnosis Consensus point-scoring system (GED-C PSS; Mehta et al., 2019) and Finnish biobank data and specimens in the automated point scoring of large populations. An indicative point-score range for Finnish GD patients was determined, but undiagnosed patients were not identified partly due to high number of high-score subjects in combination with a lack of suitable samples for diagnostics in the assessed biobank population. The current study extended the screening to another biobank and evaluated the feasibility of utilising the automated GED-C PSS in conjunction with small nucleotide polymorphism (SNP) chip genotype data from the FinnGen study of biobank sample donors in the identification of undiagnosed GD patients in Finland. Furthermore, the applicability of FFPE tissues and DNA restoration in the next-generation sequencing (NGS) of the GBA gene were tested.
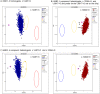
Methods: Previously diagnosed Finnish GD patients eligible to the study, and up to 45,100 sample donors in Helsinki Biobank (HBB) were point scored. The GED-C point scoring, adjusted to local data, was automated, but also partly manually verified for GD patients. The SNP chip genotype data for rare GBA variants was visually assessed. FFPE tissues of GD patients were obtained from HBB and Biobank Borealis of Northern Finland (BB).
Results: Three previously diagnosed GD patients and one patient previously treated for GD-related features were included. A genetic diagnosis was confirmed for the patient treated for GD-related features. The GED-C point score of the GD patients was 12.5-22.5 in the current study. The score in eight Finnish GD patients of the previous and the current study is thus 6-22.5 points per patient. In the automated point scoring of the HBB subpopulation (N ≈ 45,100), the overall scores ranged from 0 to 17.5, with 0.77% (346/45,100) of the subjects having ≥10 points. The analysis of SNP chip genotype data was able to identify the diagnosed GD patients, but potential undiagnosed patients with the GED-C score and/or the GBA genotype indicative of GD were not discovered. Restoration of the FFPE tissue DNA improved the quality of the GBA NGS, and pathogenic GBA variants were confirmed in five out of six unrestored and in all four restored FFPE DNA samples.
Discussion: These findings imply that the prevalence of diagnosed patients (~1:325,000) may indeed correspond the true prevalence of GD in Finland. The SNP chip genotype data is a valuable tool that complements the screening with the GED-C PSS, especially if the genotyping pipeline is tuned for rare variants. These proof-of-concept biobank tools can be adapted to other rare genetic diseases.
Keywords: BB, Biobank Borealis of Northern Finland; Biobank study; DF4/DF5, Data freeze 4/5; EHR, Electronic health record; Electronic health record data; FFPE, Formalin-fixed, paraffin embedded; GBA; GBA1/GBA, β-glucocerebrosidase gene; GD, Gaucher disease; GED-C, Gaucher Earlier Diagnosis Consensus; Gaucher disease; Gaucher earlier diagnosis consensus point-scoring system; GlcCer, β-glucosylceramide; GlcCerase, β-glucosylceramidase; GlcSph/Lyso-Gb1, β-glucosylsphingosine; HBB, Helsinki Biobank; HUH, Helsinki University Hospital; HUS, Hospital District of Helsinki and Uusimaa; ICD-10, International Statistical Classification of Diseases and Related Health Problems 10th Revision; NGS, Next-generation sequencing; OUH, Oulu University Hospital; PSS, Point-scoring system; SNP, small nucleotide polymorphism; Small nucleotide polymorphism chip genotype data; VUS, variant of uncertain significance.
© 2022 The Authors.
Conflict of interest statement
KE is employed by Takeda (Helsinki, Finland) and holds stocks/stock options in Takeda. HH, MP, SM, PB, and OC are employed by Helsinki Biobank (Helsinki, Finland) which received reimbursement from Medaffcon and Takeda, for the work done at the biobank. KU and ML are employed by Medaffcon Oy (Espoo, Finland) which received reimbursement from Takeda, for conducting the study. UWK reports consultancy fees from Medaffcon and Takeda during the study, as well as grant support, paid to her institution, from several foundations for research outside the submitted work. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Figures


Similar articles
-
Assessing the diagnostic utility of the Gaucher Earlier Diagnosis Consensus (GED-C) scoring system using real-world data.Orphanet J Rare Dis. 2024 Feb 16;19(1):71. doi: 10.1186/s13023-024-03042-y. Orphanet J Rare Dis. 2024. PMID: 38365689 Free PMC article.
-
The Gaucher earlier diagnosis consensus point-scoring system (GED-C PSS): Evaluation of a prototype in Finnish Gaucher disease patients and feasibility of screening retrospective electronic health record data for the recognition of potential undiagnosed patients in Finland.Mol Genet Metab Rep. 2021 Feb 9;27:100725. doi: 10.1016/j.ymgmr.2021.100725. eCollection 2021 Jun. Mol Genet Metab Rep. 2021. PMID: 33604241 Free PMC article.
-
Scoring system to facilitate diagnosis of Gaucher disease.Intern Med J. 2020 Dec;50(12):1538-1546. doi: 10.1111/imj.14942. Intern Med J. 2020. PMID: 33174353 Free PMC article.
-
Presenting signs and patient co-variables in Gaucher disease: outcome of the Gaucher Earlier Diagnosis Consensus (GED-C) Delphi initiative.Intern Med J. 2019 May;49(5):578-591. doi: 10.1111/imj.14156. Intern Med J. 2019. PMID: 30414226 Free PMC article. Review.
-
Glucosylsphingosine (Lyso-Gb1) as a reliable biomarker in Gaucher disease: a narrative review.Orphanet J Rare Dis. 2023 Feb 13;18(1):27. doi: 10.1186/s13023-023-02623-7. Orphanet J Rare Dis. 2023. PMID: 36782327 Free PMC article. Review.
Cited by
-
Obstacles to Early Diagnosis of Gaucher Disease.Ther Clin Risk Manag. 2025 Jan 25;21:93-101. doi: 10.2147/TCRM.S388266. eCollection 2025. Ther Clin Risk Manag. 2025. PMID: 39882275 Free PMC article. Review.
-
Assessing the diagnostic utility of the Gaucher Earlier Diagnosis Consensus (GED-C) scoring system using real-world data.Orphanet J Rare Dis. 2024 Feb 16;19(1):71. doi: 10.1186/s13023-024-03042-y. Orphanet J Rare Dis. 2024. PMID: 38365689 Free PMC article.
References
-
- Stirnemann J., Belmatoug N., Camou F., Serratrice C., Froissart R., Caillaud C., Levade T., Astudillo L., Serratrice J., Brassier A., Rose C., Billette de Villemeur T., Berger M. A review of Gaucher disease pathophysiology, clinical presentation and treatments. IJMS. 2017;18:441. doi: 10.3390/ijms18020441. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

