Geometric complement heterogeneous information and random forest for predicting lncRNA-disease associations
- PMID: 36092871
- PMCID: PMC9448985
- DOI: 10.3389/fgene.2022.995532
Geometric complement heterogeneous information and random forest for predicting lncRNA-disease associations
Abstract
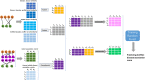
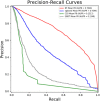
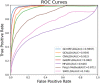
More and more evidences have showed that the unnatural expression of long non-coding RNA (lncRNA) is relevant to varieties of human diseases. Therefore, accurate identification of disease-related lncRNAs can help to understand lncRNA expression at the molecular level and to explore more effective treatments for diseases. Plenty of lncRNA-disease association prediction models have been raised but it is still a challenge to recognize unknown lncRNA-disease associations. In this work, we have proposed a computational model for predicting lncRNA-disease associations based on geometric complement heterogeneous information and random forest. Firstly, geometric complement heterogeneous information was used to integrate lncRNA-miRNA interactions and miRNA-disease associations verified by experiments. Secondly, lncRNA and disease features consisted of their respective similarity coefficients were fused into input feature space. Thirdly, an autoencoder was adopted to project raw high-dimensional features into low-dimension space to learn representation for lncRNAs and diseases. Finally, the low-dimensional lncRNA and disease features were fused into input feature space to train a random forest classifier for lncRNA-disease association prediction. Under five-fold cross-validation, the AUC (area under the receiver operating characteristic curve) is 0.9897 and the AUPR (area under the precision-recall curve) is 0.7040, indicating that the performance of our model is better than several state-of-the-art lncRNA-disease association prediction models. In addition, case studies on colon and stomach cancer indicate that our model has a good ability to predict disease-related lncRNAs.
Keywords: autoencoder; geometric complement heterogeneous information; lncRNA-disease association prediction; machine learning; random forest.
Copyright © 2022 Yao, Zhang, Zhan, Zhang, Zhan and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32. 10.1023/a:1010933404324 - DOI
LinkOut - more resources
Full Text Sources