The worldwide spread of Aedes albopictus: New insights from mitogenomes
- PMID: 36092930
- PMCID: PMC9459080
- DOI: 10.3389/fgene.2022.931163
The worldwide spread of Aedes albopictus: New insights from mitogenomes
Abstract
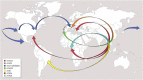
The tiger mosquito (Aedes albopictus) is one of the most invasive species in the world and a competent vector for numerous arboviruses, thus the study and monitoring of its fast worldwide spread is crucial for global public health. The small extra-nuclear and maternally-inherited mitochondrial DNA represents a key tool for reconstructing phylogenetic and phylogeographic relationships within a species, especially when analyzed at the mitogenome level. Here the mitogenome variation of 76 tiger mosquitoes, 37 of which new and collected from both wild adventive populations and laboratory strains, was investigated. This analysis significantly improved the global mtDNA phylogeny of Ae. albopictus, uncovering new branches and sub-branches within haplogroup A1, the one involved in its recent worldwide spread. Our phylogeographic approach shows that the current distribution of tiger mosquito mitogenome variation has been strongly affected by clonal and sub-clonal founder events, sometimes involving wide geographic areas, even across continents, thus shedding light on the Asian sources of worldwide adventive populations. In particular, different starting points for the two major clades within A1 are suggested, with A1a spreading mainly along temperate areas from Japanese and Chinese sources, and A1b arising and mainly diffusing in tropical areas from a South Asian source.
Keywords: Aedes albopictus spread; MtDNA variation; haplogroups; mitogenome; phylogeny; sources of adventive populations.
Copyright © 2022 Battaglia, Agostini, Moroni, Colombo, Lombardo, Rambaldi Migliore, Gabrieli, Garofalo, Gagliardi, Gomulski, Ferretti, Semino, Malacrida, Gasperi, Achilli, Torroni and Olivieri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Adhami J., Murati N. (1987). Presence du moustique Aedes albopictus en Albanie. Rev. Mjekesore 1, 13–16.
-
- Beebe N. W., Ambrose L., Hill L. A., Davis J. B., Hapgood G., Cooper R. D., et al. (2013). Tracing the tiger: Population genetics provides valuable insights into the Aedes (stegomyia) albopictus invasion of the australasian region. PLoS Negl. Trop. Dis. 7, e2361. 10.1371/journal.pntd.0002361 - DOI - PMC - PubMed
-
- Bellini R., Calvitti M., Medici A., Carrieri M., Celli G., Maini S. (2007). “Use of the sterile insect technique against Aedes albopictus in Italy: First results of a pilot trial,” in Use of the sterile insect technique against Aedes albopictus in Italy: First results of a pilot trial” in area-wide control of insect pests. Editors Vreysen M. J. B., Robinson A. S., Hendrichs J. (Dordrecht, Netherlands: Springer; ), 505–515. 10.1007/978-1-4020-6059-5_47 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

