Lack of Association between Epidermal Growth Factor or Its Receptor and Reflux Esophagitis, Barrett's Esophagus, and Esophageal Adenocarcinoma: A Case-Control Study
- PMID: 36092955
- PMCID: PMC9459439
- DOI: 10.1155/2022/8790748
Lack of Association between Epidermal Growth Factor or Its Receptor and Reflux Esophagitis, Barrett's Esophagus, and Esophageal Adenocarcinoma: A Case-Control Study
Abstract
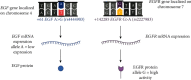
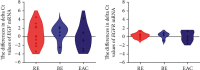
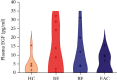
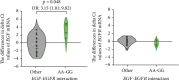
The epidermal growth factor (EGF) and its receptor (EGFR) gene-gene interactions were shown to increase the susceptibility to esophageal cancer. However, the role of the EGF/EGFR pathway in the development of gastroesophageal reflux disease (GERD) and its complications (reflux esophagitis (RE), Barrett's esophagus (BE), and esophageal adenocarcinoma (EAC)) remains unclear. This association study is aimed at investigating functional EGF and EGFR gene polymorphisms, their mRNA expression in esophageal tissues, and EGF plasma levels in relation to RE, BE, and EAC development in the Central European population. 301 patients with RE/BE/EAC (cases) as well as 98 patients with nonerosive reflux disease (NERD) and 8 healthy individuals (controls) were genotyped for +61 A>G EGF (rs4444903) and +142285 G>A EGFR (rs2227983) polymorphisms using the TaqMan quantitative polymerase chain reaction (qPCR). In random subgroups, the EGF and EGFR mRNA expressions were analyzed by reverse transcription qPCR in esophageal tissue with and without endoscopically visible pathological changes; and the EGF plasma levels were determined by enzyme-linked immunosorbent assay. None of the genotyped SNPs nor EGF-EGFR genotype interactions were associated with RE, BE, or EAC development (p > 0.05). Moreover, mRNA expression of neither EGF nor EGFR differed between samples of the esophageal tissue with and without endoscopically visible pathology (p > 0.05) nor between samples from patients with different diagnoses, i.e., RE, BE, or EAC (p > 0.05). Nevertheless, the lower EGF mRNA expression in carriers of combined genotypes AA +61 EGF (rs4444903) and GG +142285 EGFR (rs2227983; p < 0.05) suggests a possible direct/indirect effect of EGF-EGFR gene interactions on EGF gene expression. In conclusion, EGF and EGFR gene variants and their mRNA/protein expression were not associated with RE, BE or EAC development in the Central European population.
Copyright © 2022 Tereza Deissova et al.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Interaction of epidermal growth factor with COX-2 products and peroxisome proliferator-activated receptor-γ system in experimental rat Barrett's esophagus.Am J Physiol Gastrointest Liver Physiol. 2020 Mar 1;318(3):G375-G389. doi: 10.1152/ajpgi.00410.2018. Epub 2020 Jan 13. Am J Physiol Gastrointest Liver Physiol. 2020. PMID: 31928220
-
A functional epidermal growth factor (EGF) polymorphism, EGF serum levels, and esophageal adenocarcinoma risk and outcome.Clin Cancer Res. 2008 May 15;14(10):3216-22. doi: 10.1158/1078-0432.CCR-07-4932. Clin Cancer Res. 2008. PMID: 18483390 Free PMC article.
-
Epidermal growth factor A61G gene polymorphism, gastroesophageal reflux disease and esophageal adenocarcinoma risk.Carcinogenesis. 2009 Aug;30(8):1363-7. doi: 10.1093/carcin/bgp126. Epub 2009 Jun 11. Carcinogenesis. 2009. PMID: 19520791 Free PMC article.
-
Reflux injury of esophageal mucosa: experimental studies in animal models of esophagitis, Barrett's esophagus and esophageal adenocarcinoma.Dis Esophagus. 2007;20(5):372-8. doi: 10.1111/j.1442-2050.2007.00713.x. Dis Esophagus. 2007. PMID: 17760649 Review.
-
Gastroesophageal reflux and Barrett's esophagus: a pathway to esophageal adenocarcinoma.Updates Surg. 2018 Sep;70(3):339-342. doi: 10.1007/s13304-018-0564-y. Epub 2018 Jul 23. Updates Surg. 2018. PMID: 30039277 Review.
Cited by
-
Molecular Abnormalities and Carcinogenesis in Barrett's Esophagus: Implications for Cancer Treatment and Prevention.Genes (Basel). 2025 Feb 25;16(3):270. doi: 10.3390/genes16030270. Genes (Basel). 2025. PMID: 40149421 Free PMC article. Review.
References
-
- Long J. D., Orlando R. C. Nonerosive reflux disease. Minerva Gastroenterologica e Dietologica . 2007;53(2):127–141. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

