Biological effects and regulation of IGFBP5 in breast cancer
- PMID: 36093095
- PMCID: PMC9453429
- DOI: 10.3389/fendo.2022.983793
Biological effects and regulation of IGFBP5 in breast cancer
Abstract
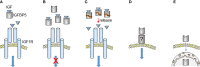
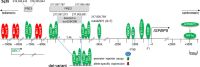
The insulin-like growth factor receptor (IGF1R) pathway plays an important role in cancer progression. In breast cancer, the IGF1R pathway is linked to estrogen-dependent signaling. Regulation of IGF1R activity is complex and involves the actions of its ligands IGF1 and IGF2 and those of IGF-binding proteins (IGFBPs). Six IGFBPs are known that share the ability to form complexes with the IGFs, by which they control the bioavailability of these ligands. Besides, each of the IGFBPs have specific features. In this review, the focus lies on the biological effects and regulation of IGFBP5 in breast cancer. In breast cancer, estrogen is a critical regulator of IGFBP5 transcription. It exerts its effect through an intergenic enhancer loop that is part of the chromosomal breast cancer susceptibility region 2q35. The biological effects of IGFBP5 depend upon the cellular context. By inhibiting or promoting IGF1R signaling, IGFBP5 can either act as a tumor suppressor or promoter. Additionally, IGFBP5 possesses IGF-independent activities, which contribute to the complexity by which IGFBP5 interferes with cancer cell behavior.
Keywords: breast cancer; breast cancer susceptibility region; estrogen; insulin-like growth factor binding protein 5; transcriptional regulation.
Copyright © 2022 Dittmer.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

